Team:CSIA SouthKorea
From 2012hs.igem.org
(Difference between revisions)
Eugeniestar (Talk | contribs) (→Simulation of the display) |
|||
| (168 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | [[Image:CSIA_SouthKorea_team.png|right|frame|OUR FANTASTIC TEAM LOGO by Aileen Shin]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Our team is consisted of four students who are fond of thinking creatively, sharing our knowledge with others and making contributions to the society. We hope that iGEM 2012 could be a great opportunity for us to get our feet wet in the field of synthetic biology and interact with many other students around the world who are also interested in this field! Since our school does not have facilities for wet lab, team CSIA_SouthKorea asked professor In-Geol Choi in Korea University for guide. Professor Choi, Instructor Hyeok-Jin Ko and Hongjae Park let us to be familiar with synthetic biology, taught us how to use basic lab facilities and helped every part in our projects :) | |
| - | + | [[Image:CSIA_SouthKorea.png|left|frame|TEAM LOGO♥]] | |
| - | + | ||
| - | + | ||
| - | + | [[Image:CSIA_SouthKorea2.png|left|frame|TEAM LOGO]] | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 53: | Line 21: | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | =Team= | ||
| + | |||
| + | : Our [[Team:CSIA_SouthKorea/team | team]] and school is introduced in this page! :) | ||
=Project= | =Project= | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | ==Abstract== | |
| - | : | + | : Based on the design of ''V.fischeri'', we placed luxR gene under luxpL promotor and placed luxI, Aiia, and GFP gene under luxpR promotor. In this V.fischeri quorum sensing system<sup>1</sup>, LuxI synthase produces an acyl-homoserine lactone (AHL), which is a small molecule diffuses extracellularly and triggers quorum sensing. When AHL binds to LuxR, it produces LuxR–AHL complex that activates luxI promoter. This also activate GFP genes, so fluorescence can be detected. AiiA 'represses'<sup>2</sup> continuing activation of luxI promotor by degrading of AHL. Therefore, fluorescence may have the cycle under right conditions. |
| + | : This network which an activator triggers its own repressor illustrates a concept of synthetic oscillator design. Further, theoretical work shows how an autoinducer leads synchronized oscillations in a single cell and a population of cells<sup>3</sup>. | ||
| + | : The team got interested in synchronized oscillator while reading Team Wageningen's 2011 project. However, we modified their model a little to increase the probability of success in experiment by using each luxpL and luxpR promoter for only one time. | ||
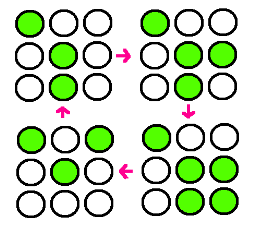
| - | + | : Our research goal is to build the circuit for synchronized oscillator and use them for display. Based on the idea that oscillatory period differs for different cell population, we plan to have a 3 x 3 array, each well consisted of different colonies. The difference in period of GFP expression will allow to display certain figure at certain time. For instance, the display could be like this: | |
| - | + | ||
| + | : [[File:gfp.jpg]] | ||
| - | + | : In the world where people suffer from energy deficiency, we expect that this technology could be applied to many different areas. Among them, we think the most successful adaptation would be as an alternative for signs, secret messages and night stand. | |
| - | + | ||
| - | = | + | ==Introduction of E.Coli display using repressilator== |
| + | : Our [[Team:CSIA_SouthKorea/introduction | introduction]] further explains the system and goals. | ||
| - | = | + | ==Mechanism of the circuit== |
| + | : This is further explanation about [[Team:CSIA_SouthKorea/mechanism | mechanism]] of the circuit and explanation about the parts we used. | ||
| - | = | + | ==Design of the circuit== |
| + | : This is the ultimate [[Team:CSIA_SouthKorea/design | design]] of the circuit and how we cloned different parts together. | ||
| - | = | + | ==Variables that determine period of the circuit== |
| - | + | ||
| + | : Since we assumed that difference of the period in each array leads to different patterns, it is important to find the [[Team:CSIA_SouthKorea/variables | variables]] which affect the period. | ||
| + | |||
| + | |||
| + | ==Simulation of the display== | ||
| + | |||
| + | : Before doing the wet lab, in order to predict the results of our experiment, we did computer [[Team:CSIA_SouthKorea/simulation | simulation]] of the light bulbs of the night stand that we are trying to make with colonies of ''E. coli''. | ||
| + | |||
| + | ==Protocols== | ||
| + | |||
| + | : You will see us conducting the experiments according to the [[Team:CSIA_SouthKorea/protocols | protocols]]. | ||
| + | |||
| + | |||
| + | ==Applications== | ||
| + | |||
| + | : The page introduces possible [[Team:CSIA_SouthKorea/applications | application]] of our unique use of synthetic oscillator. | ||
| + | |||
| + | |||
| + | ==Results & Conclusions== | ||
| + | |||
| + | : This [[Team:CSIA_SouthKorea/results | page]] shows the results and conclusions of our project so far! | ||
| + | |||
| + | |||
| + | =Outreach & Human Practice= | ||
| + | |||
| + | : [[Team:CSIA_SouthKorea/outreach | Outreach]] will lead you to a page about our introducing brochure about synthetic biology, and Synbio class in Suri Nature School. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =Brainstorming= | ||
| + | |||
| + | : Please see our [[Team:CSIA_SouthKorea/brainstorming | Brainstorming]] section. We have more than ten ideas explained! | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =Notebook= | ||
| + | |||
| + | : Please see our [[Team:CSIA_SouthKorea/Diary | Notebook]]. We have an extensive log from September 2011 on our experiments! :) | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =Safety= | ||
| + | |||
| + | : In this page, [[Team:CSIA_SouthKorea/safety | Safety]] , you will see our team CSIA_SouthKorea's lab safety practice and learnings. | ||
| + | |||
| + | |||
| + | |||
| + | =References= | ||
| + | # Danino et al, A synchronized quorum of genetic clocks”, Nature vol. 463, 326-330 (2010) | ||
| + | # Liu, D. et al. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures. Biochemistry 47, 7706–7714 (2008). | ||
| + | # Garcia-Ojalvo, J., Elowitz, M. & Strogatz, S. Modeling a synthetic multicellular clock: repressilators coupled by quorum sensing. Proc. Natl Acad. Sci. USA 101, 10955–10960 (2004). | ||
| + | # http://avena.pw.usda.gov | ||
<forum_subtle /> | <forum_subtle /> | ||
Latest revision as of 11:12, 17 June 2012
Our team is consisted of four students who are fond of thinking creatively, sharing our knowledge with others and making contributions to the society. We hope that iGEM 2012 could be a great opportunity for us to get our feet wet in the field of synthetic biology and interact with many other students around the world who are also interested in this field! Since our school does not have facilities for wet lab, team CSIA_SouthKorea asked professor In-Geol Choi in Korea University for guide. Professor Choi, Instructor Hyeok-Jin Ko and Hongjae Park let us to be familiar with synthetic biology, taught us how to use basic lab facilities and helped every part in our projects :)
Contents |
Team
- Our team and school is introduced in this page! :)
Project
Abstract
- Based on the design of V.fischeri, we placed luxR gene under luxpL promotor and placed luxI, Aiia, and GFP gene under luxpR promotor. In this V.fischeri quorum sensing system1, LuxI synthase produces an acyl-homoserine lactone (AHL), which is a small molecule diffuses extracellularly and triggers quorum sensing. When AHL binds to LuxR, it produces LuxR–AHL complex that activates luxI promoter. This also activate GFP genes, so fluorescence can be detected. AiiA 'represses'2 continuing activation of luxI promotor by degrading of AHL. Therefore, fluorescence may have the cycle under right conditions.
- This network which an activator triggers its own repressor illustrates a concept of synthetic oscillator design. Further, theoretical work shows how an autoinducer leads synchronized oscillations in a single cell and a population of cells3.
- The team got interested in synchronized oscillator while reading Team Wageningen's 2011 project. However, we modified their model a little to increase the probability of success in experiment by using each luxpL and luxpR promoter for only one time.
- Our research goal is to build the circuit for synchronized oscillator and use them for display. Based on the idea that oscillatory period differs for different cell population, we plan to have a 3 x 3 array, each well consisted of different colonies. The difference in period of GFP expression will allow to display certain figure at certain time. For instance, the display could be like this:
- In the world where people suffer from energy deficiency, we expect that this technology could be applied to many different areas. Among them, we think the most successful adaptation would be as an alternative for signs, secret messages and night stand.
Introduction of E.Coli display using repressilator
- Our introduction further explains the system and goals.
Mechanism of the circuit
- This is further explanation about mechanism of the circuit and explanation about the parts we used.
Design of the circuit
- This is the ultimate design of the circuit and how we cloned different parts together.
Variables that determine period of the circuit
- Since we assumed that difference of the period in each array leads to different patterns, it is important to find the variables which affect the period.
Simulation of the display
- Before doing the wet lab, in order to predict the results of our experiment, we did computer simulation of the light bulbs of the night stand that we are trying to make with colonies of E. coli.
Protocols
- You will see us conducting the experiments according to the protocols.
Applications
- The page introduces possible application of our unique use of synthetic oscillator.
Results & Conclusions
- This page shows the results and conclusions of our project so far!
Outreach & Human Practice
- Outreach will lead you to a page about our introducing brochure about synthetic biology, and Synbio class in Suri Nature School.
Brainstorming
- Please see our Brainstorming section. We have more than ten ideas explained!
Notebook
- Please see our Notebook. We have an extensive log from September 2011 on our experiments! :)
Safety
- In this page, Safety , you will see our team CSIA_SouthKorea's lab safety practice and learnings.
References
- Danino et al, A synchronized quorum of genetic clocks”, Nature vol. 463, 326-330 (2010)
- Liu, D. et al. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures. Biochemistry 47, 7706–7714 (2008).
- Garcia-Ojalvo, J., Elowitz, M. & Strogatz, S. Modeling a synthetic multicellular clock: repressilators coupled by quorum sensing. Proc. Natl Acad. Sci. USA 101, 10955–10960 (2004).
- http://avena.pw.usda.gov
<forum_subtle />
 "
"