Team:Tyngsboro MA Tigers/Project
From 2012hs.igem.org

Project Title PROJECT TITLE
Contents |
E. COfactory: An olfactory test for the detection of carbon monoxide
According to the National Conference of State Legislatures [1], 25 states have statutes requiring residential carbon monoxide detectors. Currently, the common household CO detectors sold on the market are only sound-based. However, as people age, their senses become less acute. We are investigating the development of a genetic circuit for an indole-free E. coli chassis that converts the odorless carbon monoxide into an indole output - the naturally produced smell of bacteria. Coupled with the common household detector, we might be able to greatly reduce the number of CO-related deaths. Our cell could help save even more lives with another sensory warning of the hidden danger.
A colorless, odorless, and soundless killer takes the lives of nearly 2,100 unsuspecting United States citizens every year; that translates to an average of three people every day. Carbon monoxide poisoning is the leading cause of unintentional poisoning deaths in the United States, and one in every 2,400 people falls victim to CO poisoning, unaware of the invisible attacker. People over the age of 65 have the highest risk of falling victim to this silent killer [2]. Only by being aware of the grave danger, and understanding the structure and function of this gas, we can create a circuit that produces a scent to CO and a warning before it's too late.
The mechanism for CO detection in our system is derived from the bacterium Rhodospirillum rubrum that naturally metabolizes CO as an energy source. This photosynthetic bacterium accomplishes this task in the following chemical reaction: CO + H2O --> CO2 + H2
The enzyme carbon monoxide dehydrogenase (CODH) is a peripheral membrane protein that carries out the primary oxidation of CO to CO2 and then passes the products through a ferredoxin-like sub-unit. R. rubrum also has a carbon monoxide-sensing protein called CooA that activates the gene expression of oxidation enzymes. In our research of R. rubrum, we found that a Turkey iGEM team in the 2010 college division developed CooA and CooA-responsive promoter BioBricks they wanted to make to transform into E. coli. They were able to successfully build these parts, and now our goal is to try and put them into our own genetic circuit that, in the presence of carbon monoxide, will output an unpleasant warning.
Human Practices
Currently, the common CO detectors on the market are only sound-based. However, as people age, their senses aren't quite what they used to be, and our cell could help save even more lives with another sensory warning of the hidden danger. Also, many work places, such as construction sites, are extremely loud, easily loud enough to drown out the little beeping of a sensor, however, a disgusting and identifiable smell permeating through the compound could again save even more lives. Coupled with the common household detector, our cell could greatly reduce the number of CO deaths per year.
Creating the Genetic Circuit
Once we had our idea solidified, the next step was coming up with a circuit to make our idea into a reality. Using the [http://www.tinkercell.com/ TinkerCell] program to create an example of the cell, we wanted to follow the KISS rule and keep things as simple as possible. The 2010 METU iGEM team created a [http://partsregistry.org/wiki/index.php?title=Part:BBa_K352014 part] that successfully detected carbon monoxide, and contained a Lacl promoter, RBS, and CooA, but from that point what the output was seemed to be fairly open for change. We wanted the output smell to be very unpleasant and trigger a negative response, and decided indole, the smell naturally made by e. coli, would be an excellent choice. We chose tnaA, the indole producing gene, as the last major step in our genetic sequence, and an [http://partsregistry.org/wiki/index.php?title=Part:BBa_J45999 odorless chassis] so that indole wouldn't be naturally produced by the detector.
Obstacles in creating the circuit were finding a tnaA producing gene (still working on that) and...
The parts chosen were all marked as 'working' in the parts registry, and most had the same assembly methods, making them compatible. Our major concerns are that we won't be able to find a tnaA gene by itself, or that the parts chosen won't work be compatible in practice.
Also, special thanks go out to our iGEM adviser, Alyssa Henning!
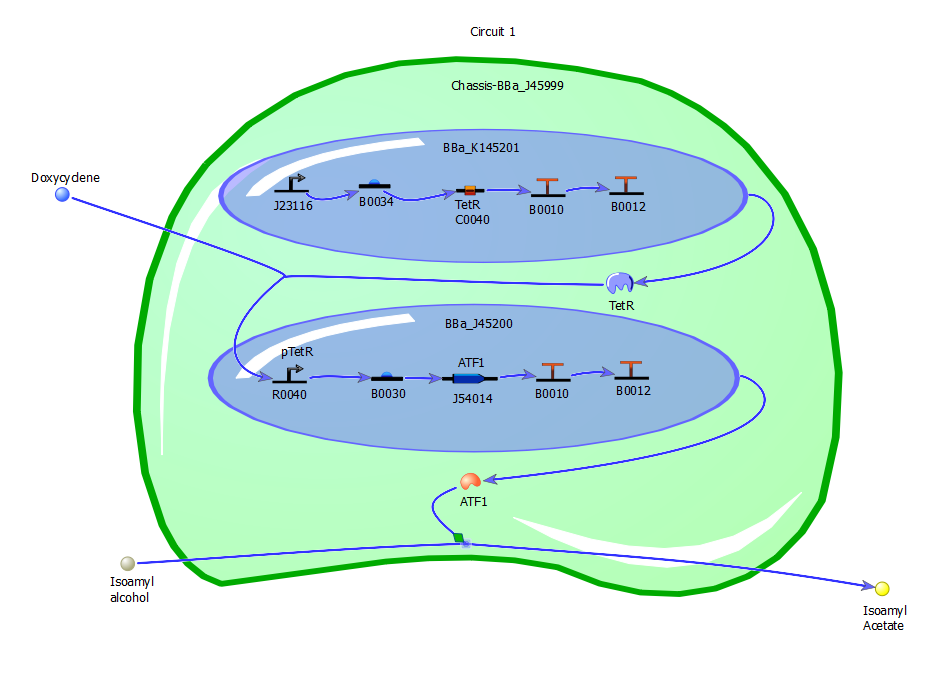
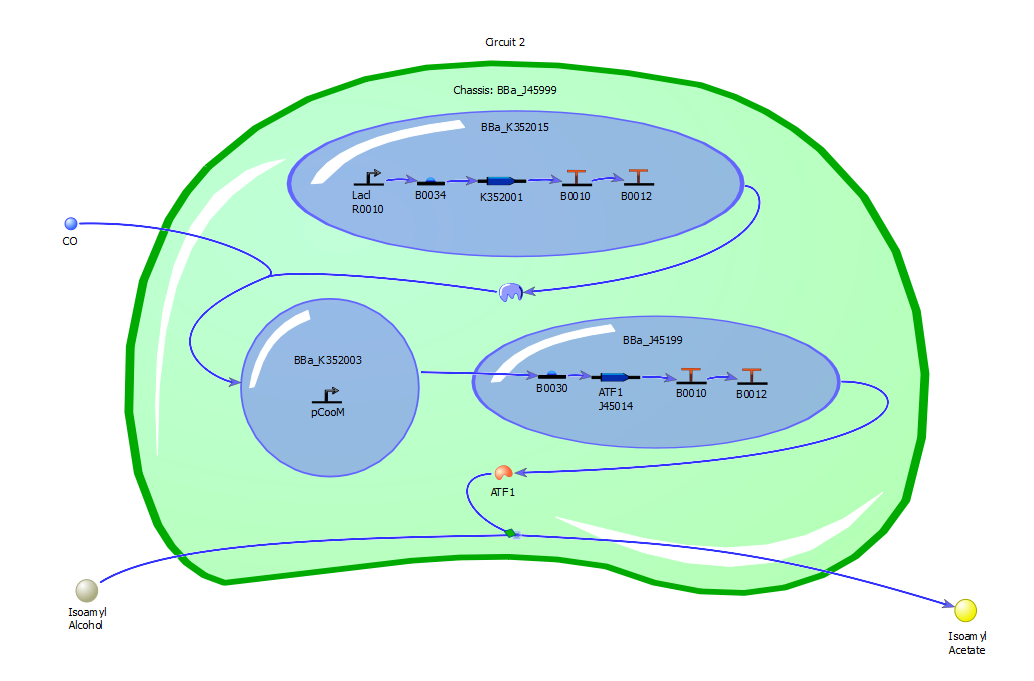
Our Circuits
Chassis (for both circuits)
[http://partsregistry.org/wiki/index.php?title=Part:BBa_J45999 BBa_J45999] – MIT’s odor free E. coli chassis.
Circuit One
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K145201 BBa_K145201] - INPUT TetR constitutive generator. TetR will repress transcription from a TetR repressible promoter. Kanamycin resistance available.
[http://partsregistry.org/wiki/index.php?title=Part:BBa_J45200 BBa_J45200] – Banana odor generator. Includes TetR repressible promoter. Ampicillin and tetracycline resistance available.
(Note: Another method of this system may be connecting the J23116 promoter from K145201 and directly attaching it to J45199, which would make the TetR promoter directly connected to the banana odor generator.)
This first circuit is our ‘proof of concept,’ designed to show that the ‘on/off’ switch can be used to control a system’s output. Without doxycycline, K145201 is allowed to run and creates TetR. J45200 has a TetR repressible promoter, meaning when TetR is present it doesn’t run and therefore doesn’t produce a banana smell. However, when K145201 is deactivated by the presence of doxycycline TetR is not produced and J45200 is allowed to run, so the isoamyl alcohol is converted into isoamyl acetate by the protein produced by J45200, and a banana smell is detected.
Circuit Two
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K352015 BBa_K352015] - CooA coupled with placI+RBS+Double Terminator – Turkey’s CO detecting part, version that includes RBS and terminating sequences. Not available.
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K352003 BBa_K352003] - pCooM Promoter from Rhodospirillum rubrum – Promoter from R. rubrum to go with K352015. Available.
[http://partsregistry.org/wiki/index.php?title=Part:BBa_J45199 BBa_J45199] – Banana odor generator, no tetR repressible promoter.
This circuit is designed to be our real carbon monoxide detector.
Protocol
We are considering the following protocols for preparing our competent cells and preforming our transformation:
[http://partsregistry.org/Help:Protocols/Competent_Cells Competent Cells]
[http://partsregistry.org/Help:Protocols/Transformation Transformation]
Here is the standard procedure we are considering for building our biobrick:
[http://partsregistry.org/Assembly:Standard_assembly Biobrick Assembly]
References
1. "Carbon Monoxide Poisoning Statistics." Christensen Law Firm. 03 Sept. 2010. Web. 1 Mar. 2012. [<http://www.utahpersonalinjurylawfirm.com/2010/09/carbon-monoxide-poisoning-statistics/>].
2. November. 2011. Web. 1 Mar. 2012. http://www.ncsl.org/issues-research/env-res/carbon-monoxide-detectors-state-statutes.aspx
3. Fox, Jeffrey D., Robert L. Kerby, Gary P. Roberts, and Paul W. Ludden. "Characterization of the CO-Induced, CO-Tolerant Hydrogenase from Rhodospirillum Rubrum and the Gene Encoding the Large Subunit of the Enzyme." Journal of Bacteriology (1996): 1515-524. American Society for Microbiology, Mar. 1996. Web. 12 Jan. 2012.
4. Mar. 2012. http://bestanimations.com/animals/mammals/cats/tigers/tigerroaringanimation-18.gif/
5. Wood, W. A., I. C. Gunsalus, and W. W. Umbreit. Function of Pyridoxal Phosphate: Resolution and Purification of the Tryptophanase Enzyme of Escherichia Coli. Publication. Ithaca: Labaratory of Bacteriology, College of Agriculture, Cornell University, 1947. Web. 22 Mar. 2012. <http://www.jbc.org/content/170/1/313.full.pdf>.
 "
"







