Team:Lethbridge Canada/Results
From 2012hs.igem.org
| Line 7: | Line 7: | ||
!align="center"|[[Image:Project.JPG|center|897px]] | !align="center"|[[Image:Project.JPG|center|897px]] | ||
|} | |} | ||
| - | |||
{| style="color:#000000;background-color:#b5cde2;" cellpadding="6" cellspacing="3" border="0" bordercolor="#00FFFF" width="65%" align="center" | {| style="color:#000000;background-color:#b5cde2;" cellpadding="6" cellspacing="3" border="0" bordercolor="#00FFFF" width="65%" align="center" | ||
Revision as of 07:23, 17 June 2012

| Home | The Team | The Project | Results | Human Practices | Notebook | Safety |
|---|
ResultsParts CreatedWe created two new parts, [http://partsregistry.org/Part:BBa_K736000 BBa_K736000] (which regulated the transcription of downstream genes) and [http://partsregistry.org/Part:BBa_K736001 BBa_K736001] (which was a TAT signal sequence fused to a red fluorescent protein, E1010). We also assembled two parts, [http://partsregistry.org/wiki/index.php?title=Part:BBa_K736002 BBa_K736002] (which tests the ability of glucose to regulate transcription and uses enhanced RFP as the reporter) and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K736003 BBa_K736003] (which tests for the ability of the TAT sequence to cause secretion of protein outside of the cell and also uses RFP as a reporter). We also created one part that coded for human insulin chain A ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K736004 BBa_K736004]) and one part that coded for human insulin chain B ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K736005 BBa_K736005]). We are also submitting the DNA for [http://partsregistry.org/Part:BBa_K331009 K331009] which was in its planning stage in the registry.
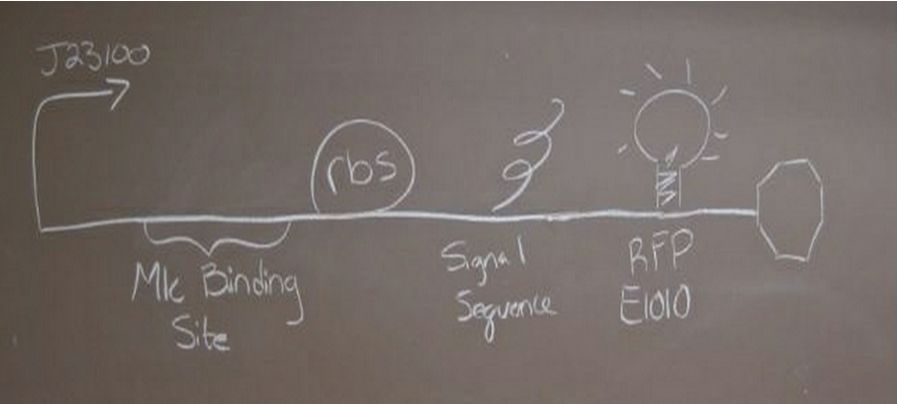
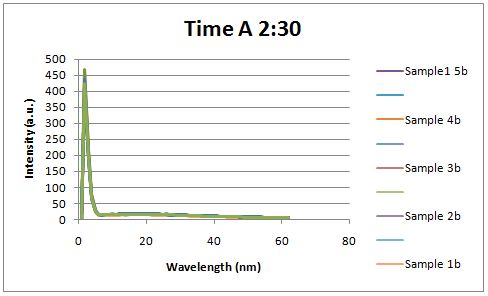
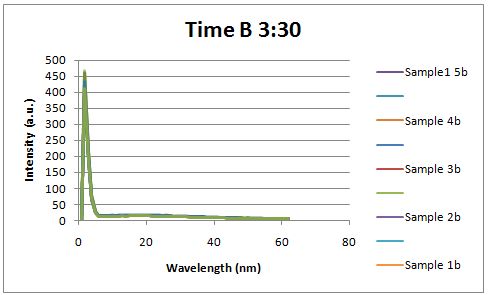
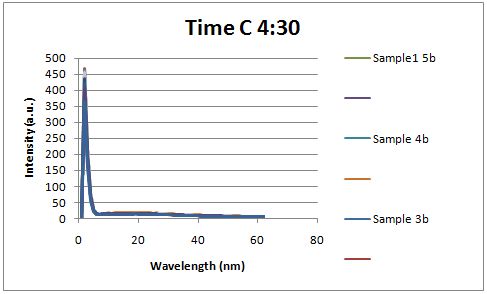
Insulin Red Fluorescent Protein Proof of PrincipleTAT signal sequence RFP with Cells Media at Different Induction Levels Preliminary results Cultures were grown containing LacI promoter attached to a twin arginine tag (TAT) signal sequence and fused to a red fluorescent protein (E1010). Samples were taken at various time points and examined for fluorescence when excited at 586nm with an expected emission at 607nm. We used a fluorescence spectrophotometer to measure the red fluorescent protein present in the supernatant after spinning the cells to a pellet. We used a constutively expressed construct expressing Red fluorescent protein( J23100 in J61002)
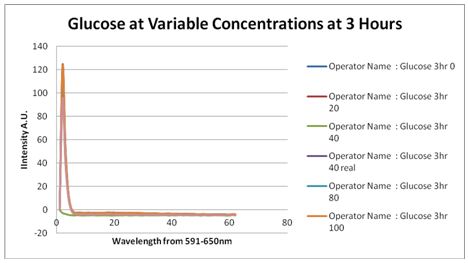
Glucose Concentration AssayPreliminary Results Cultures were grown containing constitutively expressed promoter J23100 attached to an MlC binding sight sequence and a red fluorescent protein. Samples were inoculated in varying concentrations of Glucose (0=0 mg/dl, 20= 12.5mg/dl, 40=25mg/dl, 80=50mg/dl, 160=100mg/dl, 320=200mg/dl, 1600=1000mg/dl). Samples were taken after 1.5 hour time points post inoculation and examined for fluorescents when excited at 586nm with an expected emission at 607nm. We used a fluorescents spectrophotometer to measure the red fluorescent protein present in cell lysate after spinning the cells to a pellet and re-suspending in 8M urea.
Fluorescence was measured six hours after innoculation, no emissions were observed.
|
 "
"