Team:Lethbridge Canada/Notebook
From 2012hs.igem.org
(→April) |
(→Sub-groups) |
||
| Line 113: | Line 113: | ||
'''Human Practices'''<br> | '''Human Practices'''<br> | ||
| - | ''This group is responsible for letting the | + | ''This group is responsible for letting the public know what our team is and what we are doing. This can be done creatively. The point is to raise awareness for iGEM, synthetic biology, and diabetes.'' |
Leader: Chris <br> | Leader: Chris <br> | ||
1. Shammamah <br> | 1. Shammamah <br> | ||
| Line 123: | Line 123: | ||
7.Janelle <br> | 7.Janelle <br> | ||
8. Orion <br> | 8. Orion <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
===WHMIS tests and Safety Training=== | ===WHMIS tests and Safety Training=== | ||
Revision as of 19:59, 10 June 2012

| Home | The Team | The Project | Results | Human Practices | Notebook | Safety |
|---|
NotebookMarchBrainstormingProject ideas:
Kill Switches (engineering bacteria to undergo induced or programmed cell death in order to control the organism) The chosen projectAfter dividing in to a few groups based on which project was preferred, each group made a presentation on the proposed project. In the end, we decided to go with the glucose sensing project. Project outlinePossible things to consider Glucose and insulin Immune response Kill switch Delivery system AprilSub-groupsWe divided the team into sub-groups. Each group assumed responsibility for a different aspect of the project. Wiki and Mascot Design
WHMIS tests and Safety TrainingWe are required to have safety training in order to work in the lab, and so we all wrote our WHMIS tests and did our Hazards Assesment and lab orientation! MayMay 8, 2012: Tansformation of Parts from Kit Plate and Growing Cells from Glycerol StocksTransformation of parts from kit plate: -B0034 in pSB1A2 Overnight cultures were also made of: -pMA-T K33109 (HST-1)
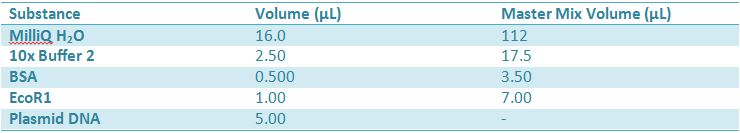
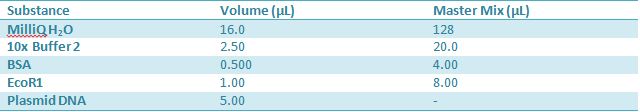
May 9, 2012: Miniprep and Restriction of PartsOvernight cultures from May 8th were miniprepped and restricted in order to determine sizes. Master mix was prepared by mixing the ingredients together in order, as shown above. 20µL worth of master mix was combined with 5µL of plasmid DNA (1-5). -pMA-T BBa_K331009 (Heat-stable toxin signal sequence) Restrictions were incubated at 37˚C for 30mins then stored at -20˚C.
-E1010 pSB2K3
May 10,2012: Miniprep and Restriction of PartsOvernight cultures from May 9th were miniprepped and restricted in order to determine sizes Master mix was prepared by mixing the ingredients together in order, as shown above. 20µL worth of master mix was combined with 5µL of plasmid DNA (1-7). -J23100 (J61002) Restrictions were incubated at 37˚C for 30min then stored in -20˚C.
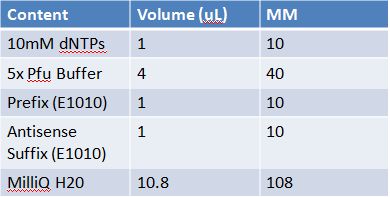
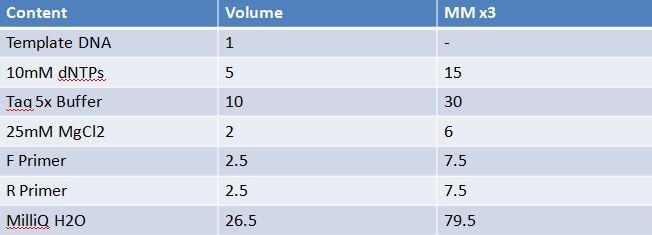
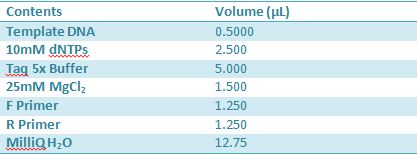
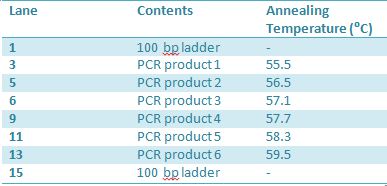
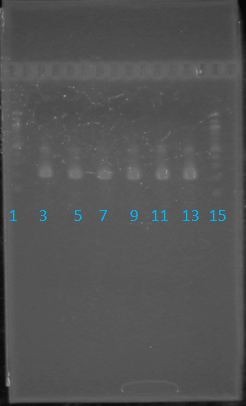
May 11th: Electrophoresis of restricted parts from May 10thPurpose: confirm existence of the following parts: -J23100 (J61002) We also grew up J04500 in pSB1AK3 for assembly PCR of RFP (E1010) into fusion standard (adding extra bp to get into fusion standard) Add 17.8 uL to 8 tubes with Pfu polymerase 0.2 uL Template DNA 2 uL Put tubes on gradient function program Pfu test with various annealing temperatures: Annealing gradient (degrees celsius):
May 12thAssembly of promoter, RBS with ss and RPF with double terminator Method: Restriction, gel extraction, & ligation Restriction: -J04500 -K331009 -E1010 (fusion) -B0015 Gel extraction We made a 1% TAE agarose gel and ran restrictions for 1 hour @ 120V Gel extracted the DNA Ligation: Ligated together J04500 & K331009 Ligated together E1010 fusion & B0015 Transformation of ligated products from May 12th
May 13thTransformation of ligated products from May 12th 2uL ligation mix to 20uL DH5ɑ, incubate on ice for 30 mins Heat shock 45sec in water bath at 42 degrees celcius Incubate on ice 5 mins Add 400uL SOC media; resuspend Shake for 1h @ 37 degrees 200uL culture onto plate Incubate @ 37 degrees for appx. 16h
May 14thPicking Cells from May 13th transformation Cells grew on 2 plates, 2 colonies were picked Picked cells were put into 5ml LB media with kanamycin Cells did not grow
May 15thCells did not grow from May 14th transformation
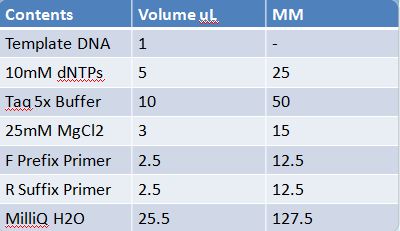
May 19th: PCR of E1010 and J23100 (Mlc primers)E1010 -Add 49.5uL to 4 PCR tubes and add 0.5uL Taq polymerase and 1uL template DNA to each -E1010 (fusion) primer annealing temp – 55.5˚C J23100: -Add 49.5 uL of MM to 4 PCR tubes and add 0.5 uL Taq polymerase to each tube -J23100-Mlc primer annealing temp- 58.0˚C -Ran 1.5% agarose TAE gel at 90V for 45 mins -E1010 expected size: 700 bp -J23100 expected size: 215 bp -Didn’t work May 20: Redo may 19 PCR of E1010 (fusion) and J23100-mlcAlterations to PCR: -2uL MgCl2 PCR cycle: -First Denaturation: 3 mins @ 94˚ -Denaturation: 15 sec @94˚ -Annealing: 30sec E1010: 54.5˚ J23100: 55.0˚ -Elongation: I min 30 sec @ 72˚ -Final elongation: 7 mins @72˚ -Add 49.5 uL of MM to 4 PCR tubes -Add 1uL template DNA -Add 0.5 uL Taq DNA polymerase to each -Didn’t work (again!)
May21st: PCR of J23100 and E1010 repeat again!
-Put 45.9 uL of MM into x PCR tubes -Add 3 uL of template DNA -PCR PFU test #4 -Ran on 1.5% agarose gel at 120V for 1hour PCR protocol: -First Denaturation: 2 mins @ 95˚ -Denaturation: 30 sec @95˚ -Annealing: 30sec E1010: 54.5˚ J23100: 55.0˚ -Elongation: I min 30 sec @ 72˚ -First elongation: 7 mins @72˚ -*changed to 30 cycles
May 22, 2012: PCR of J23100-MlcPCR Parameters: Initial denaturation 95˚C 3mins 30 cycles of: -denaturation 95˚C 30s -annealing (various) 30s -elongation 72˚C 1min
1.5% agarose gel Ran at 120v for 1 hr
May 23, 2012: Ligation of PCR products into pGEM Vector1) prefix –J23100-mlc-spe cut site (Red tubes) (sample 3 from May 22nd PCR) 2) Prefix-(fusion)-E1010-suffix (Black tubes) (sample 1 from May 23rd PCR) Contents: -2x Rapid Ligation Buffer 5µL -pGEM-Tor pGem-T Easy Vector 1µL -PCR Product 3 µL -T4 DNA Ligase 1µL -Miliq H2O 1 µL -Ligate at room temp for 1h 15 mins
May 23, 2012 Transformation of J23100-Mlc and E1010 fusionFor each ligation mix: -Added 2µL of ligation mix ( E1010 pcr + pGEM; J23100- mlc + pGEM) to 20.0µL of DH5α) -Incubated on ice for 30mins - 45secs in water bath @ 42˚C - immediately incubated on ice for 5mins -Added 400µL of SOC media; resuspended -Put in shaker for 1hr @ 37 ˚C -200µL cultured onto plate(4 plates, Amp) -Incubate @ 37 ˚C for approximately 16hrs
May 24, 2012: Transformation of PCR Products in pGEM Vectors in DH5ɑSample 3 from PCR product ligation For each ligation mix: -Added 2µL of ligation mix ( E1010 pcr + pGEM; J23100- mlc + pGEM) to 20.0µL of DH5α -Incubated on ice for 30mins - 45secs in water bath @ 42˚C - immediately incubated on ice for 5mins -Added 400µL of SOC media; resuspended -Put in shaker for 1hr @ 37 ˚C -200µL cultured onto plate(4 plates, Amp) -Incubate @ 37 ˚C for approximately 16hrs
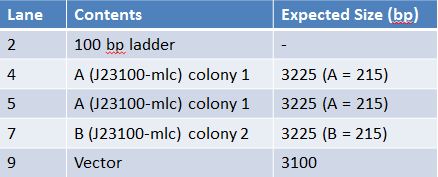
May 25, 2012: QIA Spin Miniprep of J23100-mlc (A: colony 1), J23100-mlc (B: colony 2)-Put 1.5mL of transformed cells in a microcentrifuge tube; centrifuge and repeat until cells from entire tube are in the microcentrifuge tube. -Resuspend pelleted bacterial cells in 250uL Buffer P1 -Add 250uL Buffer P2, mix by inversion -Add 350uL Buffer N3, mix by inversion -Centrifuge for 10 mins @ 13000 rpm -Put supernatant into the QIAprep spin column -Centrifuge for 60s at 14000 rpm, discard flow through -Add 750uL buffer PE and centrifuge for 60s at 14000 rpm -Centrifuge again to get rid of remaining buffer -Add 50uL of Buffer EB to spin columns, let stand for I min, centrifuge for 1 min at 14000 rpm
May 29, 2012: Transformation of E1010 fusion in pGEM in DH5ɑFor each ligation mix: -Added 2µL of ligation mix ( E1010 pcr + pGEM; J23100- mlc + pGEM) to 20.0µL of DH5α -Incubated on ice for 30mins - 45secs in water bath @ 42˚C - immediately incubated on ice for 5mins -Added 400µL of SOC media; resuspended -Put in shaker for 1hr @ 37 ˚C -200µL cultured onto plate(4 plates, Amp) -Incubate @ 37 ˚C for approximately 16hrs
May 31, 2012: Restriction ofE1010 psB2k3, E1010 fusion pGEM, J20100-mlc pGEM, CK331009 psEk3, E1010 p5B2k3
JuneJune 1, 2012: Restriction of J23100-Mlc, K093005, B0014 (samples A and B), J04500, K331009 (samples A and B), E1010 Fusion (samples A and B)
|
 "
"