Team:Lethbridge Canada/Notebook
From 2012hs.igem.org
Erin.kelly (Talk | contribs) (→Notebook) |
|||
| Line 33: | Line 33: | ||
<i> Brainstorming Project Ideas</i> | <i> Brainstorming Project Ideas</i> | ||
* <b>Pests</b> | * <b>Pests</b> | ||
| - | **Bacterial Pest attractor (engineer bacteria to produce a substance, such as a pheromone or smell, that attracts and kills pests such as insects) | + | **Bacterial Pest attractor (engineer bacteria to produce a substance, such as a pheromone or smell, that attracts and kills pests such as insects). |
| - | **Natural pesticides (engineer bacteria to produce a substance that repels or kills pests, such as those that harm crops, that can later be implemented into plants) <br> <br> | + | **Natural pesticides (engineer bacteria to produce a substance that repels or kills pests, such as those that harm crops, that can later be implemented into plants). <br> <br> |
* <b>Water/Environment</b> | * <b>Water/Environment</b> | ||
| - | **Desalination of water (engineering bacteria to get rid of salt in salt water in order to make drinkable water from sea water) | + | **Desalination of water (engineering bacteria to get rid of salt in salt water in order to make drinkable water from sea water). |
**Getting rid of estrogen mimicking compounds in water (engineering bacteria to degrade them or sequester them) | **Getting rid of estrogen mimicking compounds in water (engineering bacteria to degrade them or sequester them) | ||
| - | **Waste treatment (engineering bacteria that can be integrated into waste water treatment) | + | **Waste treatment (engineering bacteria that can be integrated into waste water treatment). |
| - | **CFCs (engineer bacteria to produce metabolites that break down chlorofluorocarbons—compounds that contribute to the degradation of the ozone layer)<br><br> | + | **CFCs (engineer bacteria to produce metabolites that break down chlorofluorocarbons—compounds that contribute to the degradation of the ozone layer).<br><br> |
* <b>Health </b> | * <b>Health </b> | ||
| - | **Stomach ulcers (creating a medication containing engineered bacteria to specifically target and kill Helicobacter pylori – the organism that causes stomach ulcers) | + | **Stomach ulcers (creating a medication containing engineered bacteria to specifically target and kill Helicobacter pylori – the organism that causes stomach ulcers). |
| - | **Diabetes (engineered bacteria as Islet cells to produce insulin) | + | **Diabetes (engineered bacteria as Islet cells to produce insulin). |
| - | **Allergies/Immune system (engineer bacteria to produce antihistamines or alter epitopes) | + | **Allergies/Immune system (engineer bacteria to produce antihistamines or alter epitopes). |
| - | **Bone density (engineer bacteria to produce and secrete calcium and other compounds to help heal broken bones or to prevent osteoporosis) <br><br> | + | **Bone density (engineer bacteria to produce and secrete calcium and other compounds to help heal broken bones or to prevent osteoporosis). <br><br> |
* <b>Process Improvement </b> | * <b>Process Improvement </b> | ||
| - | **Oil fractionation catalyst (engineering bacteria to improve the separation of crude oil into valuable fraction and waste fraction) | + | **Oil fractionation catalyst (engineering bacteria to improve the separation of crude oil into valuable fraction and waste fraction). |
| - | **Nitrate fixation (engineer bacteria to improve nitrogen fixation so not as much fertilizer is needed) <br><br> | + | **Nitrate fixation (engineer bacteria to improve nitrogen fixation so not as much fertilizer is needed). <br><br> |
* <b>Kill Switches </b> | * <b>Kill Switches </b> | ||
| - | **Engineering bacteria to undergo induced or programmed cell death in order to control the organism) <br><br> | + | **Engineering bacteria to undergo induced or programmed cell death in order to control the organism). <br><br> |
===The chosen project=== | ===The chosen project=== | ||
| Line 60: | Line 60: | ||
===Project outline=== | ===Project outline=== | ||
| - | <i>Possible things to consider</i> | + | <i>Possible things to consider:</i> |
* <b>Glucose and insulin </b> | * <b>Glucose and insulin </b> | ||
| Line 70: | Line 70: | ||
* <b>Kill switch</b> | * <b>Kill switch</b> | ||
| - | ** Control and | + | ** Control and <br><br> |
* <b>Delivery system </b> | * <b>Delivery system </b> | ||
| Line 84: | Line 84: | ||
'''Wiki and Mascot Design'''<br> | '''Wiki and Mascot Design'''<br> | ||
| - | <Blockquote>''This group is responsible to obtaining and uploading all necessary information to the wiki. This includes team pictures and biographies, experiments and results, and the projects of the other sub-groups. This will be a priority over mascot design'' | + | <Blockquote>''This group is responsible to obtaining and uploading all necessary information to the wiki. This includes team pictures and biographies, experiments and results, and the projects of the other sub-groups. This will be a priority over mascot design.'' |
<Blockquote>Leader: Erin Kelly<br> | <Blockquote>Leader: Erin Kelly<br> | ||
1. Erin Kwan<br> | 1. Erin Kwan<br> | ||
| Line 100: | Line 100: | ||
'''Biosafety'''<br> | '''Biosafety'''<br> | ||
| - | <Blockquote>''This group must document the safety protocols that the Lethbridge team practices in the lab'' | + | <Blockquote>''This group must document the safety protocols that the Lethbridge team practices in the lab.'' |
<Blockquote>Leader: Marissa Guzzi<br> | <Blockquote>Leader: Marissa Guzzi<br> | ||
1. Branden Black<br> | 1. Branden Black<br> | ||
| Line 1,409: | Line 1,409: | ||
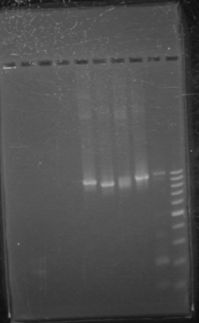
[[Image:June14gel.JPG|center|199px]] | [[Image:June14gel.JPG|center|199px]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
='''Information Sheet'''= | ='''Information Sheet'''= | ||
Revision as of 20:37, 16 June 2012

| Home | The Team | The Project | Results | Human Practices | Notebook | Safety |
|---|
NotebookMarchBrainstorming Project Ideas
The chosen project
Project outlinePossible things to consider:
AprilSub-groupsWe divided the team into sub-groups. Each group assumed responsibility for a different aspect of the project. Wiki and Mascot Design This group is responsible to obtaining and uploading all necessary information to the wiki. This includes team pictures and biographies, experiments and results, and the projects of the other sub-groups. This will be a priority over mascot design.Leader: Erin Kelly
This group must document the safety protocols that the Lethbridge team practices in the lab.Leader: Marissa Guzzi
This group is responsible for letting the public know what our team is and what we are doing. This can be done creatively. The point is to raise awareness for iGEM, synthetic biology, and diabetes.Leader: Chris Isaac WHMIS tests and Safety TrainingWe are required to have safety training in order to work in the lab, and so we all wrote our WHMIS tests and did our Hazards Assesment and lab orientation! MayMay 8, 2012: Tansformation of Parts from Kit Plate and Growing Cells from Glycerol StocksTransformation of parts from kit plate:
Overnight cultures were also made of:
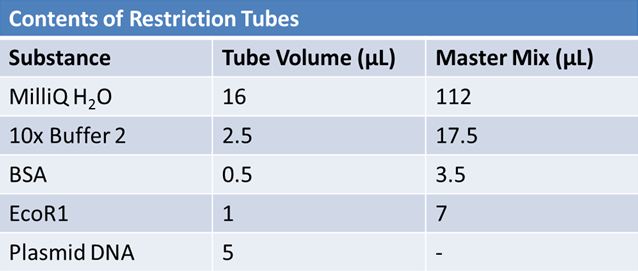
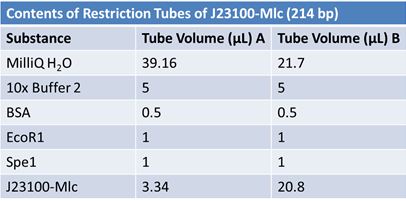
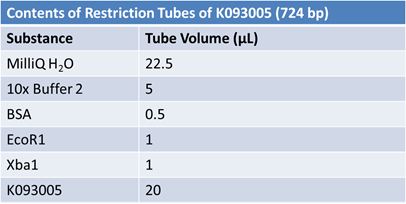
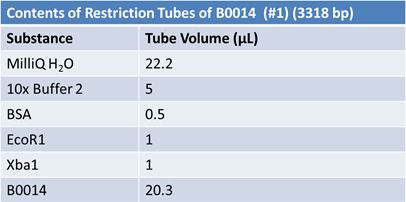
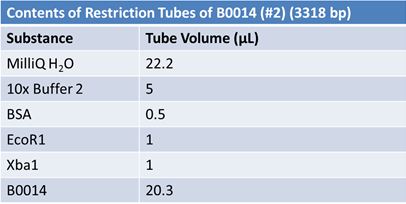
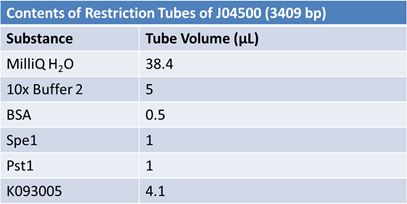
May 9, 2012: Miniprep and Restriction of PartsOvernight cultures from May 8th were miniprepped and restricted in order to determine sizes.
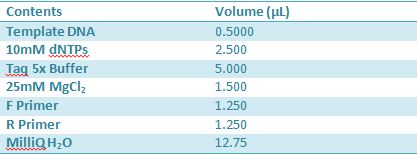
Master mix was prepared by mixing the ingredients together in order, as shown above. 20µL worth of master mix was combined with 5µL of plasmid DNA:
Restrictions were incubated at 37˚C for 30mins then stored at -20˚C.
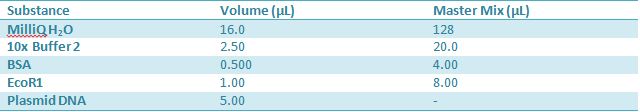
May 10, 2012: Miniprep and Restriction of PartsOvernight cultures from May 9th were miniprepped and restricted in order to determine sizes
Master mix was prepared by mixing the ingredients together in order, as shown above. 20µL worth of master mix was combined with 5µL of plasmid DNA (1-7).
Restrictions were incubated at 37˚C for 30min then stored in -20˚C. May 11th: Electrophoresis of restricted parts from May 10th
Purpose: confirm existence of the following parts:
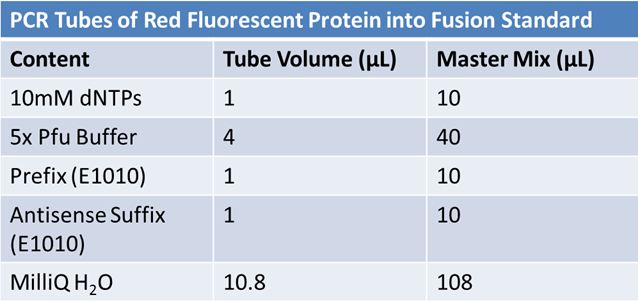
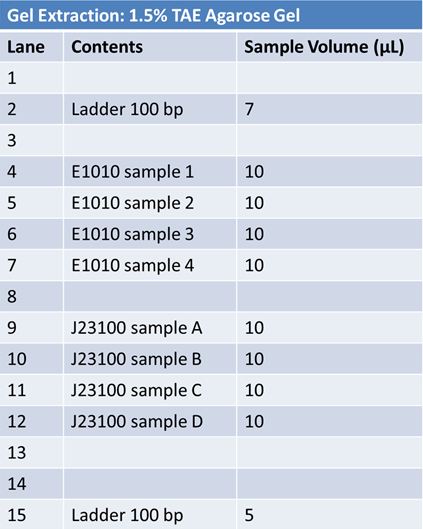
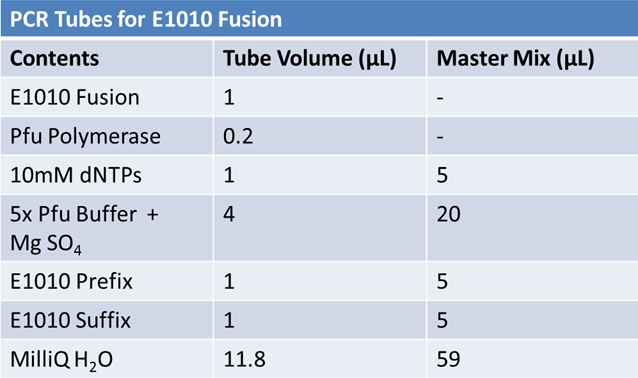
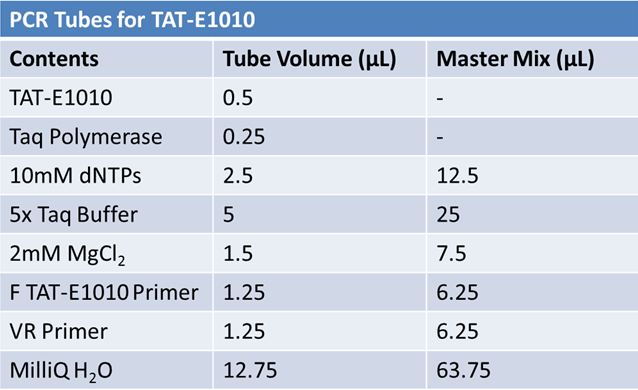
PCR of RFP (E1010) into fusion standard (adding extra bp to get into fusion standard) Add 17.8 uL to 8 tubes with: May 12th: Assembly of promoter & RBS (J04500) with signal sequence (K331009) and RPF (E1010 fusion) with double terminator (B0015)Method: Restriction, gel extraction, and ligation Restriction:
We made a 1% TAE agarose gel and ran restrictions for 1 hour @ 120V We then gel extracted the DNA Ligation:
May 13th Transformation of ligated products: J04500 + K331009 and E1010 (fusion) + B0015Transformation: 2uL of each ligation mix was added to 20uL DH5ɑ cells, then incubated on ice for 30 mins
May 14th: Picking Cells from May 13th transformation
Cells did not grow
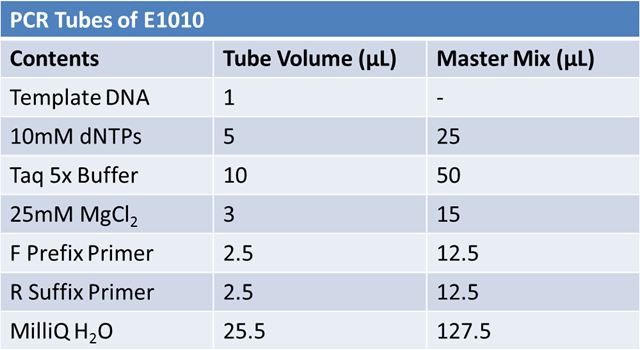
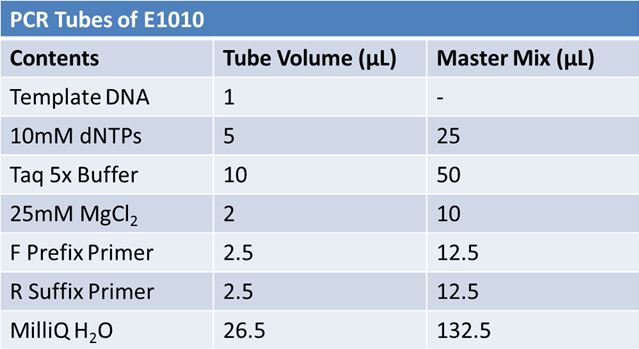
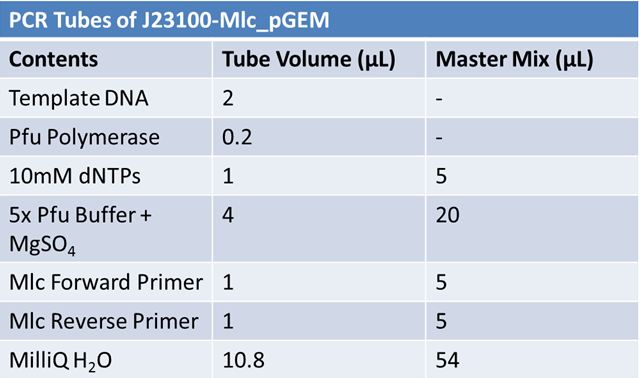
May 19th: PCR of E1010 and J23100 (Mlc primers)E1010:
Ran 1.5% agarose TAE gel at 90V for 45 mins
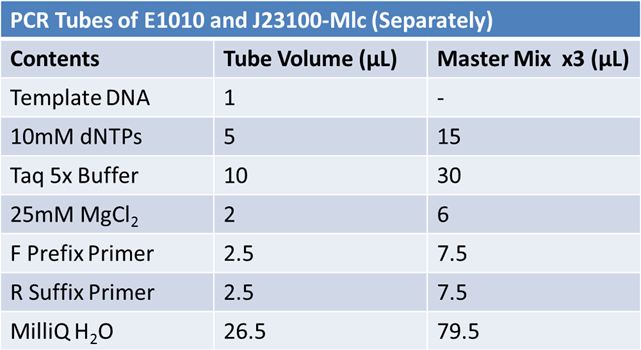
Did not work May 20: Re-do May 19 PCR of E1010 (fusion) and J23100-mlcAlterations to PCR:
PCR cycle:
May 21st: PCR of J23100 and E1010 repeat again!
For both J20100 and E1010*:
PCR protocol:
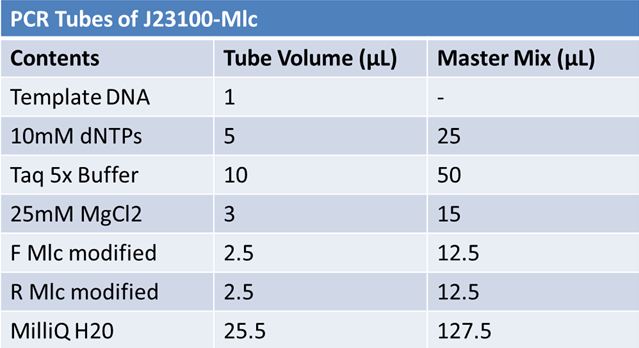
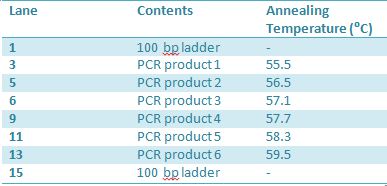
May 22, 2012: PCR of J23100-MlcPCR Parameters: Initial denaturation 95˚C 3mins 30 cycles of: *denaturation 95˚C 30s *annealing (various) 30s *elongation 72˚C 1min
1.5% agarose gel Ran at 120v for 1 hr
May 23, 2012: Ligation of PCR products into pGEM Vector1) prefix –J23100-mlc-spe cut site (Red tubes) (sample 3 from May 22nd PCR) 2) Prefix-(fusion)-E1010-suffix (Black tubes) (sample 1 from May 23rd PCR) Contents:
May 23, 2012 Transformation of J23100-Mlc and E1010 fusionFor each ligation mix:
May 23rd continuedMake: 1.5% agarose gel -0.45 agarose M -30 mL TAE (1x)
May 24, 2012: Transformation of PCR Products in pGEM Vectors in DH5ɑSample 3 from PCR product ligation For each ligation mix:
May 24 picking colonies of J20100-mlc and E1010 in DH5ɑFor each sample:
May 25, 2012: QIA Spin Miniprep of J23100-mlc (A: colony 1), J23100-mlc (B: colony 2)
May 26 gel of Miniprep samples
May 29, 2012: Transformation of E1010 fusion in pGEM in DH5ɑFor each ligation mix:
May 30 QIA spin MiniprepDNA:
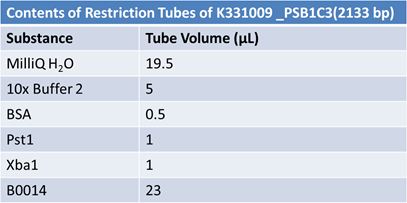
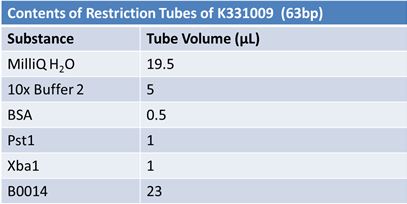
JuneJune 1, 2012: Restriction of J23100-Mlc, K093005, B0014 (samples 1 and 2), K331009_PSB1C3, K331009, E1010 Fusion (samples 1 and 2), J04500
Gel Extraction
June 2, 2012: Miniprep of June 1 Cultures
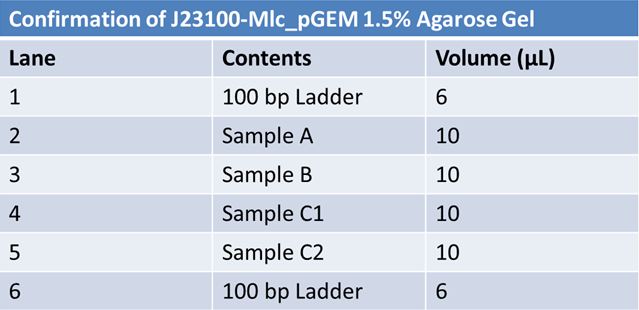
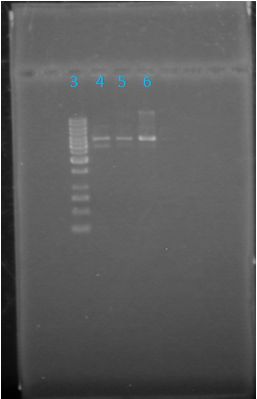
results? June 3, 2012: Confirmaiton of Parts
What worked:
June 4, 2012: PCR to Confirm J23100-Mlc in pGEM
Transformation of TAT-E1010_pGEM (x2) and J23100-Mlc_pGEM (x1) into DH5αFor each ligation mix:
June 5, 2012: Transformation of J06702 (mCherry RFP) into DH5α
Gel Extraction of June 4 RestrictionsJune 6, 2012: Gel of J23100-Mlc_pGEM Restriction (June 4)
Miniprep of J06702
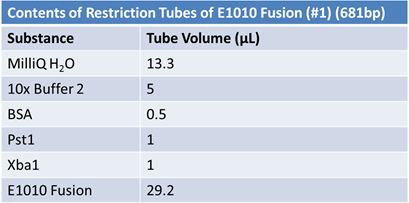
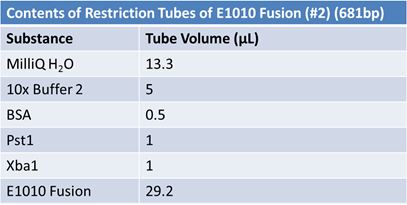
PCR of E1010 Fusion (to confirm part) and TAT-E1010 (for insertion into pGEM)
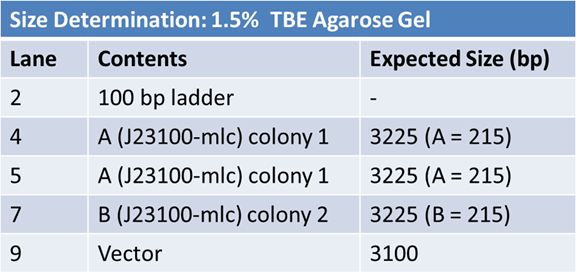
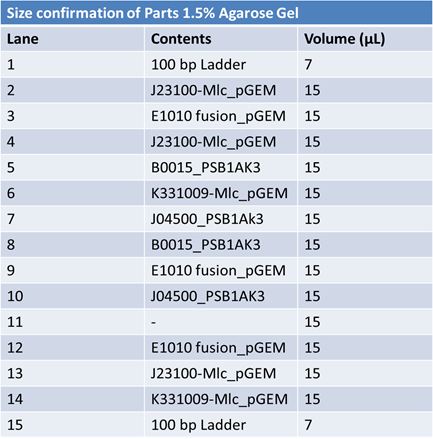
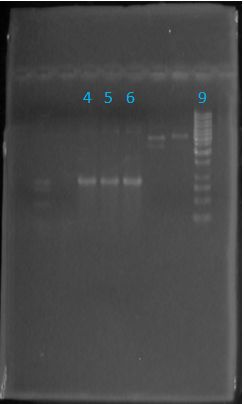
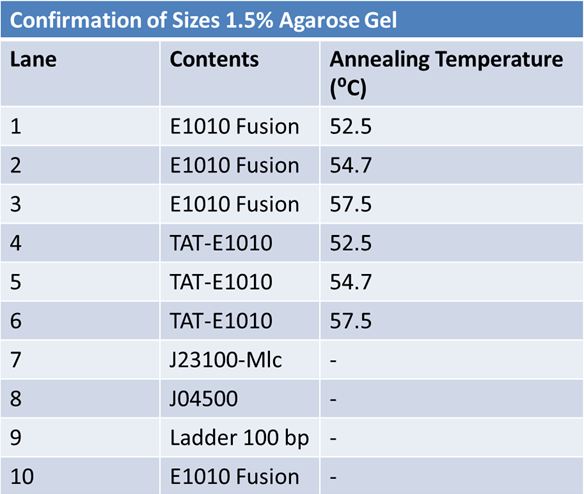
Size Confirmation Gel of E1010 fusion, TAT-E1010, J23100-Mlc, J04500
June 7th 2012
June 9th 2012
June 10th 2012
June 11th 2012
June 12th 2012
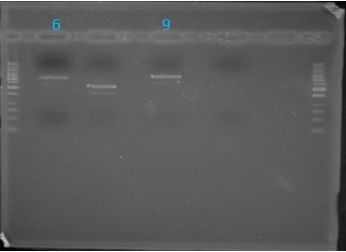
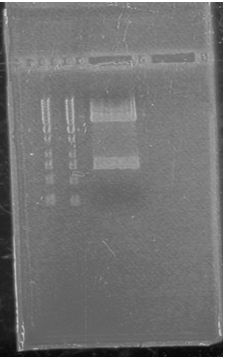
June 14th 2012Gel: Information SheetProtocols
|
 "
"