Team:Tyngsboro MA Tigers/Results
From 2012hs.igem.org
(→Circuit 1) |
m (→Circuit 1) |
||
| Line 283: | Line 283: | ||
| - | == <B>Circuit | + | == <B>Circuit 2</B> == |
If Circuit 2 was built, the following procedure would be employed: | If Circuit 2 was built, the following procedure would be employed: | ||
Revision as of 16:33, 15 June 2012

Contents |
Results
Abstract
In the United States alone, carbon monoxide is responsible for over 2,100 deaths every year, with our only current defense being the warning sounds of carbon monoxide alarms. Carbon monoxide (CO) cannot be seen or smelled, so our project idea was to develop a biological alternative to the typical carbon monoxide detector to produce a warning smell, rather than a sound. Our circuit consists of the CO transcriptional activator CooA. In the presence of a CO inducer and an isoamyl alcohol precursor, the characteristic smell of bananas will be produced. Our team has developed a proof of concept circuit using the banana odor generating device as a reporter and the Tetracycline repressor as a constitutive generator to activate the transcription of ATF1 in an indole-free chassis. This circuit will replace CO gas with doxycycline as an inducer and isoamyl alcohol as a precursor to generate the banana smell.
pUC 19 Transformation
The goal: Perform a transformation using pUC 19, a plasmid with high transformation efficiency, to test the quality of our protocol before using it for our experimental transformation.
The hypothesis: If the pUC 19 transformed successfully into E. coli, then only those cells will grow on ampicillin plates because they contain an ampicillin resistance chain.
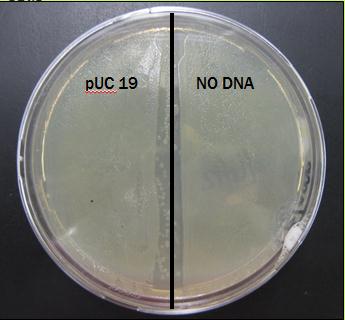
The results: The plate was covered in bacteria on both pUC 19 side and NO DNA side. Results for this preliminary test are presently inconclusive.
Expected Results
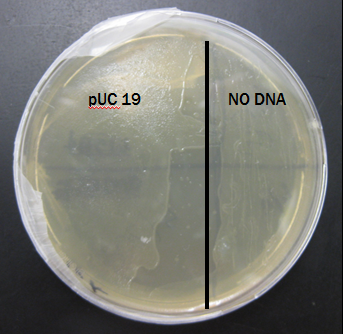
This plate shows the expected results for the transformation:
The pUC 19 side has bacteria growth and the No DNA side has no growth.
pUC 19 Transformation
Materials
Streaking from agar stabs:
70% ethanol
paper towels
marker/Sharpie
Agar Stab: - BBa_J45999 (kit)
Agar Stab: NEB10 beta strain (kit)
(6) inoculating loops (kit)
(2) LB agar plates – Amp/Kan (kit)
(1) SOB/LB agar plate (kit)
Growing up cell cultures:
(2) 14ml culture tubes (kit)
10ml of LB broth - Amp/Kan (kit)
(2) inoculating loops (kit)
Agar plate: Part A – BBa_J45999
rotator/shaker
Transformation(s)
ice
container for ice
timer
NEB10 cells (transformation 1)
Transformation 1 cells (transformation 2)
(3) 2.0ml microcentrifuge tubes (kit)
CaCl2 solution (kit)
(3) inoculating loops (kit)
SOC media (kit)
(3) LB agar plates (kit)
glass beads (kit)
waterbath
thermometer
Protocol
Part One
1. Label 2 small eppendorf tubes with “Puc 19” and “No DNA.”
2. Pipet 200 ul of CaCl2 transformation solution into each eppendorf tube and then place them on ice.
3. Use a sterile inoculating loop to gently scrape up one entire patch of the indole free cells and then swirl the cells into the “Puc 19” tube of cold CaCl2. Repeat the step for the “No DNA” tube of cold CaCl2.
4. Gently flick and invert the eppendorf tube to re-suspend the cells.
5. Keep competent cells on ice while you prepare the DNA for transformation.
Part Two
1. Retrieve the Puc 19 sample.
2. Add 2 ul of pUC 19 into the tube labeled “pUC 19”. Flick to mix the tube and return it to ice. Do not add pUC 19 to the tube labeled “No DNA."
3. Let the DNA and the cells sit on ice for 5 minutes.
4. While the cells are incubating, take a marker and label the bottom of the LB and ampicillin agar petri dish. Draw a line through the middle and label one side “Puc 19” and the other side “No DNA”, then add the date.
5. Heat shock both samples by placing the tubes at 42°C for 90 seconds exactly.
6. At the end of the 90 seconds, move the tubes to a rack at room temperature.
7. Add .5 ml of room temperature LB to the tubes. Close the caps and invert the tubes to mix the contents.
8. Using a sterile spreader, spread 250 ul of the “Puc 19” solution onto the designated half of the petri dish.
9. Using a new sterile spreader, spread 250 ul of the “No DNA” solution onto the designated half of the petri dish.
10. Cover the plate and set it aside for a minute, then turn the plate over.
11. Incubate the petri dish upside down overnight at 37°C.
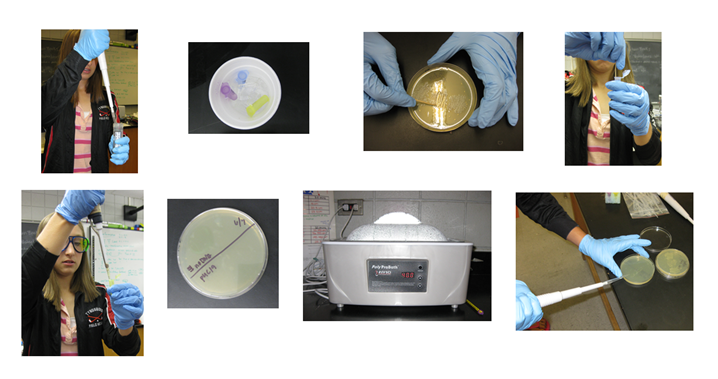
Pictures from performing different steps of the pUC19 transformation
Step 1: Pipeting CaCl2 (transformation solution) into both test tubes.
Step 2: Keeping the samples on ice.
Step 3: Scraping a patch of indole free cells.
Step 4: Adding the cells to the samples.
Step 5: Re-suspending the cells in the sample.
Step 6: Labeling the agar plate with a pUC 19 and No DNA side.
Step 7: Heat shocking the samples for 90 seconds at 42 degrees C.
Step 8: Spreading the samples onto the plates.
Results From First Trial
The plate was covered in bacteria on both sides after just 24 hours of incubation.
Results From Second Trial
The plate was also covered in bacteria on both sides after just 24 hours of incubation.
If colonies form, we will use the following procedure to determine the transformation efficiency of the plasmid. Determining Transformation Efficiency
Circuit 1
Once the transformation efficiency has been determined for pUC19, the transformation protocol will be repeated for BBa_J4500 (on ampicillin backbone). The transformants of this process will be used in a second transformation for BBa_K145201 (on kanamycin backbone). Colonies that grow on kan/amp agar plates will be selected for testing our proof of concept.
Circuit 2
If Circuit 2 was built, the following procedure would be employed:
3A Assembly Protocol
Refer to http://partsregistry.org/Help:3A_Assembly_Kit
 "
"