Team:CIDEB-UANL Mexico/Project
From 2012hs.igem.org
| (One intermediate revision not shown) | |||
| Line 456: | Line 456: | ||
That is why we decide that it would be useful to create a biosensor with bacteria to measure heavy metals concentration in samples. To test the biosensor we will use Arabinose. The reason of using Arabinose instead of heavy metals is for safety reasons because work with such materials will be a high risk to our health. Thus we first construct the model with Arabinose to see if it really works and if it does, then other biosensor which could detect the concentration of heavy metals could be constructed.</p> | That is why we decide that it would be useful to create a biosensor with bacteria to measure heavy metals concentration in samples. To test the biosensor we will use Arabinose. The reason of using Arabinose instead of heavy metals is for safety reasons because work with such materials will be a high risk to our health. Thus we first construct the model with Arabinose to see if it really works and if it does, then other biosensor which could detect the concentration of heavy metals could be constructed.</p> | ||
<p class="Estilo2">Circuit</p> | <p class="Estilo2">Circuit</p> | ||
| - | < | + | <p><img src="http://farm6.staticflickr.com/5335/7376114310_a4b1ef4d7a.jpg" width="600" height="300" alt="genes"></p> |
<p class="Estilo2"> </p> | <p class="Estilo2"> </p> | ||
<p class="Estilo2">Stand by </p> | <p class="Estilo2">Stand by </p> | ||
| Line 618: | Line 618: | ||
<p>In order to build the circuit we followed some standard protocols. In a general way we will explain how to build the circuit.</p> | <p>In order to build the circuit we followed some standard protocols. In a general way we will explain how to build the circuit.</p> | ||
| - | + | ||
<ol> | <ol> | ||
<li>Localize the DNAs that will be used, in the plates sent from the MIT and hydrate them. </li> | <li>Localize the DNAs that will be used, in the plates sent from the MIT and hydrate them. </li> | ||
| Line 627: | Line 627: | ||
</ol> | </ol> | ||
<p> </p> | <p> </p> | ||
| + | <p align="center"><img src="https://static.igem.org/mediawiki/2012hs/e/e3/Blue001c2.png" width="140" height="170"> </p> | ||
| + | <p align="center"><a href="http://openwetware.org/wiki/Image:3AAssembly.gif">http://openwetware.org/wiki/Image:3AAssembly.gif</a> </p> | ||
| + | <p>The part 3-12M is in the right side and to be cut XbaI and PstI are needed.</p> | ||
| + | <p align="center"><img src="https://static.igem.org/mediawiki/2012hs/0/04/Green001c.png" width="140" height="170"> </p> | ||
| + | <p align="center"><a href="http://openwetware.org/wiki/Image:3AAssembly.gif">http://openwetware.org/wiki/Image:3AAssembly.gif</a></p> | ||
| + | <p>If we cut a vector the side to be cut is with EcoRI and PstI enzymes.</p> | ||
| + | <p align="center"><img src="https://static.igem.org/mediawiki/2012hs/4/4b/Red001c2.png" width="140" height="170"> </p> | ||
| + | <p align="center"><a href="http://openwetware.org/wiki/Image:3AAssembly.gif">http://openwetware.org/wiki/Image:3AAssembly.gif</a> </p> | ||
| + | <p>In this way, when the 2 parts that were removed are inserted in a new vector the part 1 (the left one) will attach to the EcoRI side and the part 2 (the right one) will be attached in the PstI side. XbaI and SpeI take the place in the way that they finish in the initial order but instead of having one biobrick inside, there are 2 biobricks attached. The enzymes will have this order:</p> | ||
| + | <p align="center"><img src="https://static.igem.org/mediawiki/2012hs/2/29/Et2001.png" width="600" height="100"> </p> | ||
| + | <p align="center"><img src="http://openwetware.org/images/2/21/3AAssembly.gif" width="450" height="285"> </p> | ||
| + | <p> | ||
| + | <ol> | ||
| + | </p> | ||
| + | <ol> | ||
| + | <li>After cutting the parts we attach the two parts joining them together. For example the parts 1-6M and 3-12M are cut. Then 1-6M has ampiciline resistance and the part 3-12M has ampiciline and kanamicine resistance. We have to use a vector with a different resistance to see if the bacteria have the complete new vector when tested. Following this factor is how we chose the vector that we will use and in this case we chose tetracycline and the vector with this resistance is the PSB1T3 (1-7A).</li> | ||
| + | <li>Once the parts are together they are transformed in bacteria.</li> | ||
| + | <li> | ||
| + | <p>After that we culture in tubes with broth medium and the process is repeated until all the circuit is complete.</p> | ||
| + | </li> | ||
| + | </ol> | ||
<p class="Estilo2"> </p> | <p class="Estilo2"> </p> | ||
| + | <p align="left" class="Estilo3"> </p> | ||
| + | <p align="left" class="Estilo3"> </p> | ||
| + | <p align="left" class="Estilo3"> </p> | ||
<p> </p> | <p> </p> | ||
</div> | </div> | ||
Latest revision as of 03:55, 16 June 2012


Project
Abstract
Nowadays due to the contamination caused by pollutants such as heavy metals, it is important the implementation of simple and complete detection methods. This project aims to create a biosensor that may detect the presence of heavy metals in water. In order to construct the biosensor, a genetic circuit was built into E. coli. The circuit represents a model, which works with Arabinose. It is a semi-quantitative biosensor. The circuit has 3 parts: High concentration, Low concentration and Stand-by state. Each part of the circuit has a different response: when there is a high concentration of Arabinose the bacteria shows a yellow fluorescent color; when the concentration is low it uses a sensitivity tunner that increases the response from the same amount of Arabinose, which makes the bacteria to appear in a red fluorescence; and when there is no Arabinose in the sample, the bacteria shows a green fluorescence.
General Idea
The heavy metals which are found in the crust of the Earth and they bring us both benefits and potential damage for humanity. The various damages that they can cause are reflected especially in our health and in the environment. For this reason we are searching for an easy way to determine the presence of heavy metals in the environment by this manner we can stop harmful situations for our ambience now and in the future.
That is why we decide that it would be useful to create a biosensor with bacteria to measure heavy metals concentration in samples. To test the biosensor we will use Arabinose. The reason of using Arabinose instead of heavy metals is for safety reasons because work with such materials will be a high risk to our health. Thus we first construct the model with Arabinose to see if it really works and if it does, then other biosensor which could detect the concentration of heavy metals could be constructed.
Circuit
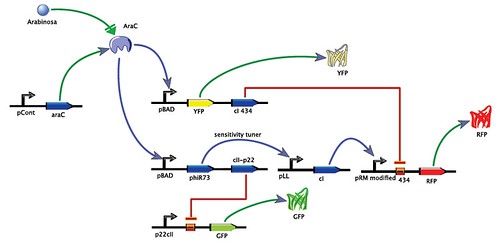
Stand by

The first part is called Stand-by. It is in a turned on all the time and it indicates that there is no presence of Arabinose in the sample. Thus we need a constitutive promoter that is always on and in this case we have the part R0053 that corresponds to the promoter p22-cll which position in the DNA plates sent from the MIT is 1-6M. We have to observe a response to know if the promoter is always working. In this case, a reporter will be used. This reporter is a GFP protein that emits green color fluorescence. The following part is a RBS followed by a reporter. In this case, we found a biobrick, K081012 with a location in plates 3-12M, that includes a RBS, a reporter, and a terminator, all together in a vector. What we have to do is to join the constitutive promoter 1-6M with the biobricks composed of 3 parts, the 3-12M.
High Concentration

In the High concentration section, we use the pBAD promoter which works with the AraC and acts as a repressor: when there is no Arabinose it represses the DNA transcription (..?) and when Arabinose is present it keeps working the pBAD. When Arabinose enters into the bacteria it joins with the AraC and these 2 mlecules joined are the ones that will be quantified. This promoter is found in a biobrick, I0500, joined with an AraC part. This biobrick has a location 1-14N in the plates. The following part is a RBS, but it has to be compatible with the circuit. We found 3 RBS compatible and we chose the B0034 which is the most recommended of the 3 in the Registry of Standard Biological Parts (http://partsregistry.org/Main_Page) and It has a location 1-2M in the plates. In this section, Also we need a response that indicates the presence of the Arabinose in high concentration, and this response will be through the YFP reporter. This part is the E0032 with a location 1-6E. Then we need another RBS and another part that will repress the section of Low concentration because Low and High concentration work with the same promoter, as we can see in the first image of the whole circuit. The repressor will act when the response of the amount of Arabinose measured is bigger than the response of the low concentration. In this way only the part of High concentration will keep on. In this part we use a biobrick which has the 3 parts: a RBS, a repressor, and a terminator. This is the part p0152 with a location 1-10E. This is the last part of the High concentration section.
Low Concentration

The parts that compose the Low concentration section are: the promoter pBAD I0500 (1-14N) which is the same of the High concentration, the part I746352 (2-12G) which has a RBS with a phiR73 and it is joined with the p0153 (1-10G). The amount of Arabinose is measured by the pBAD and then the response is increased by the sensitivity tuner. The phiR73 (that is in the 2-12G) receives the amount measured by the pBAD and it sends the signal to the pLL wich is a promoter I746365 (2-14A) which is joined with the part p0151 (1-10C). The p0151 has a RBS and the cI that multiplies the response received and it sends it to the part I12006 (2-11J). Also it has a terminator. Then the part 2-11J is the promoter that receives the response increased from the cI. It is joined with the part K081014 (3-12O) which has a RBS, the RFP that will give the red fluorescence response and a double terminator. The promoter receives the response increased and with this final response the red color appears. In this final part is where the high concentration’s repressor acts. The part 1-10G has a repressor which turns off the stand-by section of the circuit.
List of parts
In the following table we can see a list of the parts used in the Project. This list includes the parts already explained and also the vectors used in the project.
Parts |
Número |
Plate |
Well |
Plasmid |
Resistance |
pConst+RBS+araC |
I0500 |
1 |
14N |
pSB2K3 |
K/K |
RBS (elowitz) |
B0034 |
1 |
2M |
pSB1A2 |
A/A |
Promotor p22 cll |
R0053 |
1 |
6M |
pSB1A2 |
A/A |
YFP |
E0032 |
1 |
6E |
pSB1A2 |
A/A |
rbs + cl434 + doble terminador |
P0152 |
1 |
10E |
pSB1A2 |
A/A |
rbs + cL+ doble terminador |
P0151 |
1 |
10C |
pSB1A2 |
A/A |
rbs + c22 + terminador |
p0153 |
1 |
10G |
pSB1A2 |
A/A |
RBS 1 + CDS |
I746352 |
2 |
12G |
pSB1A2 |
A/A |
Promoto prm |
I12006 |
2 |
11J |
pSB2K3 |
K/K |
pLL promoter |
I746365 |
2 |
14A |
pSB1A2 |
A/A |
rbs + GFP + terminador |
K081012 |
3 |
12M |
pSB1AK3 |
AK/AK |
rbs + mRFP1 + terminador |
K081014 |
3 |
12O |
pSB1AK3 |
AK/AK |
pSB1T3 |
pSB1T3 |
1 |
7A |
T |
|
pSB1C3 |
pSB1C3 |
1 |
3A |
C |
|
pSB1AT3 |
pSB1AT3 |
1 |
13A |
AT |
|
pSB1K3 |
pSB1K3 |
1 |
5A |
K |
|
pSB1A3 |
pSB1A3 |
1 |
1G |
AT |
In order to build the circuit we followed some standard protocols. In a general way we will explain how to build the circuit.
- Localize the DNAs that will be used, in the plates sent from the MIT and hydrate them.
- Transform the DNAs into the E. Colli bacteria.
- Culture the bacteria into tubes and store some as reserves.
- The other tubes are used to do MiniPreparation of plasmid DNA in order to extract DNA from the bacteria and check their quality by running them in an agarose gel.
- Cut the parts that are inside of the vector with restriction enzymes type II. This enzymes cut in a specific place.

http://openwetware.org/wiki/Image:3AAssembly.gif
The part 3-12M is in the right side and to be cut XbaI and PstI are needed.

http://openwetware.org/wiki/Image:3AAssembly.gif
If we cut a vector the side to be cut is with EcoRI and PstI enzymes.

http://openwetware.org/wiki/Image:3AAssembly.gif
In this way, when the 2 parts that were removed are inserted in a new vector the part 1 (the left one) will attach to the EcoRI side and the part 2 (the right one) will be attached in the PstI side. XbaI and SpeI take the place in the way that they finish in the initial order but instead of having one biobrick inside, there are 2 biobricks attached. The enzymes will have this order:


- After cutting the parts we attach the two parts joining them together. For example the parts 1-6M and 3-12M are cut. Then 1-6M has ampiciline resistance and the part 3-12M has ampiciline and kanamicine resistance. We have to use a vector with a different resistance to see if the bacteria have the complete new vector when tested. Following this factor is how we chose the vector that we will use and in this case we chose tetracycline and the vector with this resistance is the PSB1T3 (1-7A).
- Once the parts are together they are transformed in bacteria.
-
After that we culture in tubes with broth medium and the process is repeated until all the circuit is complete.
Latest News
March 25, 2012
Headline
Back in the early nineties (yeah, i'm old) i was tripping ballz on acid one night with some friends.
class="date"> March 26, 2012
Headline II
At some point one of us got the brilliant idea to test out our clairvoyant abilities under the influence and we set up a nifty experiment where one of us would take a random card out of a playing deck and would try to 'send' the card telepathically to one of the others. When me and my best friend at the time were up, I ended up calling the exact card three times in a row.
March 27, 2012
Headline III
Pretty much left the room speechless. The weird thing is, when we talked about it later on we both sort of knew beforehand we could do this and couldn't stop smiling during the whole ordeal.
 "
"