Team:Tyngsboro MA Tigers/Results
From 2012hs.igem.org

Results
Abstract
In the United States alone, carbon monoxide is responsible for over 2,100 deaths every year, with our only current defense being the warning sounds of carbon monoxide alarms. Carbon monoxide (CO) cannot be seen or smelled, so our project idea was to develop a biological alternative to the typical carbon monoxide detector to produce a warning smell, rather than a sound. Our circuit consists of the CO transcriptional activator CooA. In the presence of a CO inducer and an isoamyl alcohol precursor, the characteristic smell of bananas will be produced. Our team has developed a proof of concept circuit using the banana odor generating device as a reporter and the Tetracycline repressor as a constitutive generator to activate the transcription of ATF1 in an indole-free chassis. This circuit will replace CO gas with doxycycline as an inducer and isoamyl alcohol as a precursor to generate the banana smell.
pUC 19 Transformation
The goal: Perform a transformation using pUC 19, a plasmid with high transformation efficiency, to test the quality of our protocol before using it for our experimental transformation.
The hypothesis: If the pUC 19 transformed successfully into E. coli, then only those cells will grow on ampicillin plates because they contain an ampicillin resistance chain.
The results: The plate was covered in bacteria on both pUC 19 side and NO DNA side. Results for this preliminary test are presently inconclusive.
Expected Results
This plate shows the expected results for the transformation:
The pUC 19 side has bacteria growth and the No DNA side has no growth.
pUC 19 Transformation
Materials
Protocol
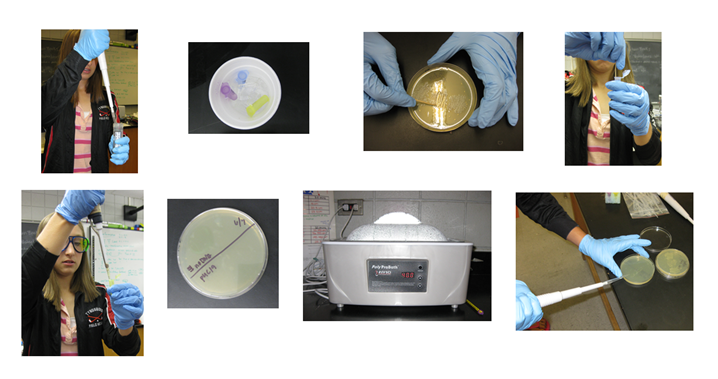
Pictures from performing different steps of the pUC19 transformation
Step 1: Pipeting CaCl2 (transformation solution) into both test tubes.
Step 2: Keeping the samples on ice.
Step 3: Scraping a patch of indole free cells.
Step 4: Adding the cells to the samples.
Step 5: Re-suspending the cells in the sample.
Step 6: Labeling the agar plate with a pUC 19 and No DNA side.
Step 7: Heat shocking the samples for 90 seconds at 42 degrees C.
Step 8: Spreading the samples onto the plates.
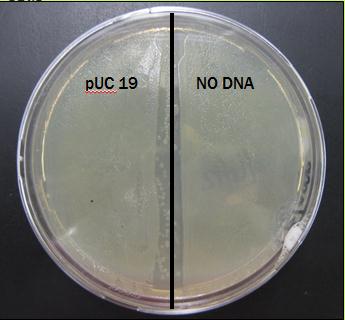
Results From First Trial
The plate was covered in bacteria on both sides after just 24 hours of incubation.
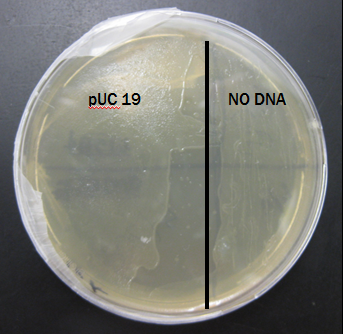
Results From Second Trial
The plate was also covered in bacteria on both sides after just 24 hours of incubation.
If colonies form, we will use the following procedure to determine the transformation efficiency of the plasmid. Determining Transformation Efficiency
Once the transformation efficiency has been determined, the transformation protocol will be repeated for BBa_J4500 (on ampicillin backbone). The transformants of this process will be used in a second transformation for BBa_K145201 (on kanamycin backbone). Colonies that grow on kan/amp agar plates will be selected for testing the system.
3A Assembly Protocol
 "
"