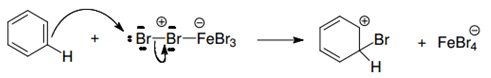Team:PrepaTec GarzaSadaMx/Protocols
From 2012hs.igem.org
(→Protocols) |
(→Protocols) |
||
| (18 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
<FONT COLOR="#ffffff"> | <FONT COLOR="#ffffff"> | ||
| - | '''Naphthalene Dioxygenase''' | + | ===='''Naphthalene Dioxygenase'''==== |
This enzyme is obtained from the cells of the Pseudomonas sp. which has a melting point of 55°C. One of the early uses of the enzyme was made during the process of cloning gens which were implicated in the degradation of naphthalene. In this process the fragments that were cloned of the plasmid nah express themselves while in contact with E. coli creating blue colonies. The enzyme also works as a catalyst, transforming indole into indigo, from which we obtain the tyrian purple. | This enzyme is obtained from the cells of the Pseudomonas sp. which has a melting point of 55°C. One of the early uses of the enzyme was made during the process of cloning gens which were implicated in the degradation of naphthalene. In this process the fragments that were cloned of the plasmid nah express themselves while in contact with E. coli creating blue colonies. The enzyme also works as a catalyst, transforming indole into indigo, from which we obtain the tyrian purple. | ||
| - | + | ||
| + | |||
| + | [[File:naphthalene_protocols.jpg|center|Naphtalene Dioxygenase]] | ||
| + | |||
What makes the indole change to indigo is an oxidation of the tryptophan that is cloned and expressed on the E. coli. The tryptophan or indole is an aromatic hydrocarbon, that when in contact with oxygen, changes to cis- dihydrodiol which makes up indigo. The oxidation is caused by combined activities of the trytophanase and napthalene diosygenase, as shown on the reaction. | What makes the indole change to indigo is an oxidation of the tryptophan that is cloned and expressed on the E. coli. The tryptophan or indole is an aromatic hydrocarbon, that when in contact with oxygen, changes to cis- dihydrodiol which makes up indigo. The oxidation is caused by combined activities of the trytophanase and napthalene diosygenase, as shown on the reaction. | ||
The optimum ph of the naphthalene dioxygenase in the Pseudomas sp. is of 7.5, meaning that the ph is almost neutral. The optimum temperature of the enzyme is 30°C. The pressure that is exerted on the enzyme is between a reduced pressure and atmospheric pressure, which is the normal pressure we feel. | The optimum ph of the naphthalene dioxygenase in the Pseudomas sp. is of 7.5, meaning that the ph is almost neutral. The optimum temperature of the enzyme is 30°C. The pressure that is exerted on the enzyme is between a reduced pressure and atmospheric pressure, which is the normal pressure we feel. | ||
| - | '''Competent cells''' | + | |
| + | ===='''Competent cells'''==== | ||
| - | Summary: | + | <ins>Summary:</ins> |
| - | + | ||
| + | |||
| + | As shown in the paper "Plasmid DNA purification and Agarose gel electrophoresis" written by several researchers, when you’re planning on taking a cell and make it recombinant, you need to make it competent. A competent cell is when you modified it in certain ways so that its membrane creates pores, to let DNA molecules pass through it and insert the plasmid on the cell. For this experiment, what we did was making E. Coli competent. In order to do this, first we need to prepare a Calcium chloride solution and do the next steps. The procurement of competent cells on E. Coli consists on concentrating a culture picked up on a mid-low growth logarithmic phase, and by successive washings in a cold solution (4°C) of Calcium chloride (CaCl2). Then mix the cells with the DNA. When the mixture is ready, you summit it 90 seconds to 42°C. This treating modifies the E.Coli membrane and increases its permeability to the DNA. This helps the plasmids enter the cell as a double strand molecule, while the linear DNA that enters the same way into the cell, are degraded by the Cytoplasmic nucleases (RecBCD). | ||
| + | |||
| - | Materials: | + | <ins>Materials:</ins> |
*Pipette | *Pipette | ||
| Line 34: | Line 41: | ||
| - | Protocol Competent cells: | + | <ins>Protocol Competent cells:</ins> |
| - | As shown in the paper made by Anh-Hue T. Tu | + | As shown in the paper made by Anh-Hue T. Tu , to make competent cells you should perform the following steps: |
#Inoculate 5 mL of LB media with E. Coli and grow overnight at 37°C on an incubator. | #Inoculate 5 mL of LB media with E. Coli and grow overnight at 37°C on an incubator. | ||
| Line 54: | Line 61: | ||
| - | '''Transformation of cells | + | ===='''Transformation of cells'''==== |
| - | Summary: | + | <ins>Summary:</ins> |
| + | |||
| + | The transformation is a process by which cells (in this case E. coli) are able to identify DNA present in the surrounding area. In order for this to take place, the E. coli has to be in a competent state.1 Some bacteria are naturally in this state, however, this is no the case of the E. coli, thus this state is achieved by following the previous process. In this state, the bacteria presents some alterations in its cellular wall and membrane, which now will allow the entrance of nucleic acids to the cell.1 The entrance of foreign DNA in the form of a plasmid –either in its circular form or in its supercoiled form- to the cell is what’s called transformation. 1 | ||
| + | Here's an animation, made by McGraw Hill, of the process in general [http://highered.mcgraw-hill.com/sites/0072556781/student_view0/chapter14/animation_quiz_2.html]. | ||
| - | |||
| + | [[File:transformation_protocols.jpg|center|Transformation]] | ||
| + | |||
| + | |||
After the cell is transformed, it has to go through the step of selection. This final step is very important, because after doing this, it’s possible that some of the bacteria didn’t take up the plasmid and therefore, in order to identify the transformed bacteria, the DNA in the plasmid has a marker.1 The marker consist in making the transformed cells resistant to the selective media. In this experiment, the media where the E. coli cells are going to be tested will contain ampicillin, and consequently only the cells with the plasmid –which will make them ampicillin resistant- are going to survive.1 By doing this, we ensure that the cells that we harvest are the ones that contain the mentioned plasmid, as they were able to develop in the selective media.1 | After the cell is transformed, it has to go through the step of selection. This final step is very important, because after doing this, it’s possible that some of the bacteria didn’t take up the plasmid and therefore, in order to identify the transformed bacteria, the DNA in the plasmid has a marker.1 The marker consist in making the transformed cells resistant to the selective media. In this experiment, the media where the E. coli cells are going to be tested will contain ampicillin, and consequently only the cells with the plasmid –which will make them ampicillin resistant- are going to survive.1 By doing this, we ensure that the cells that we harvest are the ones that contain the mentioned plasmid, as they were able to develop in the selective media.1 | ||
| - | Materials: | + | <ins>Materials:</ins> |
*Luria broth (LB) | *Luria broth (LB) | ||
| Line 89: | Line 101: | ||
*Roller drum in a 37°C incubator | *Roller drum in a 37°C incubator | ||
| - | Protocol: | + | <ins>Protocol:</ins> |
#Add 10 to 40 ng (10 to 25 ml volume) of DNA to 250 ml of competent cells. Concentrated stock DNA can be diluted using sterile 1x TE buffer. This can be done to prevent a decrease in efficiency resulting from high concentration of DNA. Besides the DNA tube, additional tubes should be set up as listed below: | #Add 10 to 40 ng (10 to 25 ml volume) of DNA to 250 ml of competent cells. Concentrated stock DNA can be diluted using sterile 1x TE buffer. This can be done to prevent a decrease in efficiency resulting from high concentration of DNA. Besides the DNA tube, additional tubes should be set up as listed below: | ||
| Line 105: | Line 117: | ||
| + | |||
| + | ===='''Protocol electrophoresis'''==== | ||
| + | |||
| + | |||
| + | <ins>Summary:</ins> | ||
| + | In order to recover plasmids from E. Coli, you need to put it through a process known as gel electrophoresis. This technique consists on summit the DNA molecule mixture in an agarose gel with pores, and then apply an electric charge. The DNA molecules are attracted to the positive pole due to their negative charge caused by the phosphate group. The larger molecules will move slower through the pores because of their size, while the smaller molecules will move faster through the gel. When the time has gone for the molecules to separate (Around an hour and a half), the gel is submerge on an ethidium bromide solution, which has an affinity for the DNA and its intercalated in-between the base pairs and its fluorescent when its bond to it. When you illuminate the solution with UV light, you can identify the fluorescent bands which correspond to the molecules of DNA. Then, you choose the DNA molecule you need to purify and with a knife or something sharp, you cut the square where the molecule is located and take it away from the gel. | ||
| + | |||
| + | <table cellpadding="5" cellspacing="0" border="0"> | ||
| + | <tr bgcolor="#1b2c8a"> | ||
| + | <td> | ||
| + | [[File:electro_protocols.jpg|center|Electrophoresis gel under UV light]] | ||
| + | </td> | ||
| + | <td> | ||
| + | <FONT COLOR="#ffffff"> | ||
| + | [[File:F18.gif|center|Electrophoresis gel]] | ||
| + | |||
| + | [http://www.phschool.com/science/biology_place/biocoach/red/gel.html Link of previous animation] | ||
| + | </FONT COLOR> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | <ins>Materials:</ins> | ||
| + | *Agarose | ||
| + | *Buffer TBE | ||
| + | *Sybr Green | ||
| + | *Microwave | ||
| + | *Electrophoreticbed | ||
| + | *Comb | ||
| + | *DNA ladder | ||
| + | *DNA simple with the desire gene | ||
| + | *Pipette | ||
| + | *UV light transiluminator | ||
| + | *Knife | ||
| + | *Power font | ||
| + | |||
| + | |||
| + | <ins>Protocol:</ins> | ||
| + | #Manufacture the agarose gel at 1%: | ||
| + | **On a flask, mix 1 g of agarose with 100 mL of the buffer TBE. | ||
| + | **Heat the mixture on a microwave till its melt and the agarose is completely dissolved. | ||
| + | **Add sybr green (Fluorescence) to the mixture of agarose and TBE. The fluorescence will help locate the DNA molecule of interest. | ||
| + | **Let the gel cool down till the temperature decreases to 50°C or 60°C. When it reaches that temperature, pour it on an electrophoretic bed. Place the comb to form the holes on the gel. On each hole, 25 μL can be pour. | ||
| + | **Without moving, let the mixture gellify at room temperature. | ||
| + | #Once the agarose gel is ready, carefully remove the comb. | ||
| + | #Take a DNA Ladder and mix it with a loaded dye (This dye help us see the movement of the DNA molecules when the electrophoresis has started). | ||
| + | #Once it has the dye, put the DNA ladder on a hole in the gel. It’s a control sample to measure the DNA molecules which have the desired length. | ||
| + | #Now, take the sample of DNA (25 μL) from which it will be obtained the plasmid and mixed it with a loaded dye. Put it in another one of the holes made by the comb in the gel. Repeat this process with another sample of DNA. | ||
| + | #Once the gel is loaded with the DNA, submit it to an electric field of 10 volts, overnight. The negative pole will be located on the side of the holes, and the positive pole on the opposite side. The molecules will move from left to right, due to the fact that DNA is charged negatively, going to the opposite pole, the positive. | ||
| + | #In the morning, check the electrophoresis with UV light transiluminator, locating the desired DNA molecule. | ||
| + | #When located cut the part of the gel where it’s located. | ||
| + | #Repeat steps 1 to 8, but now with an agarose gel at 3%. | ||
| + | |||
| + | |||
| + | [[File:electro2_protocols.jpg|center|Materials needed for electrophoresis]] | ||
| + | |||
| + | |||
| + | ===='''Miniprep'''==== | ||
| + | |||
| + | |||
| + | <ins>Summary:</ins> | ||
| + | |||
| + | In order to get the plasmid DNA out of the bacteria we used the miniprep technique, which uses different solutions and a centrifuge to separate the components inside the bacterial cells. A centrifuge is essentially a device that spins contained materials extremely fast; its units are revolutions per minute (rpm). The contents inside the centrifuge become separated from one another based on their densities: denser substances will go down to the bottom of the container, while light substances and liquids go to the top. After the centrifugation, a pipette is used to remove the liquid part of the substance and thus separate it from the solid part or parts. The miniprep technique essentially takes advantage of the centrifugation technique, using solutions (usually alkaline ones) to further separate the products of the previous centrifugation. For example, the original substance is separated using centrifugation, and say, the part of interest left is in solid state. So the next step in the miniprep protocol is add a certain solution to further separate, chemically, the substance, and then to physically separate it using centrifugation. The product of this second centrifugation will be further separated, depending on its composition, by a different solution, then centrifugated again and so on, until the final or desired substance is isolated. In this case the desired substance is the bacterial plasmids, and to chemically and physically separate it is the goal of the whole miniprep technique. It is important to note that the whole process of miniprep is done using buffers or buffered solutions, because of the importance of keeping the bacteria in controlled conditions (sudden changes of pH can kill the bacteria). | ||
| + | |||
| + | |||
| + | <ins>Materials:</ins> | ||
| + | |||
| + | *Pipette 2 ul | ||
| + | *Miniprep iGEM kit | ||
| + | *Lysogenic Broth Medium | ||
| + | *Microcentrifuge | ||
| + | *RNase | ||
| + | *Lytic enzyme buffer | ||
| + | *Wash buffer | ||
| + | *PE buffer | ||
| + | *Recuperation columns | ||
| + | *Desired bacteria | ||
| + | |||
| + | |||
| + | <ins>Protocol:</ins> | ||
| + | |||
| + | #Pipet 1 mL of bacteria into the Lysogeny Broth medium (LB). Spin them down in a microcentrifuge for 1 minute at 10,000 rpm to separate the bacteria from the solution. Repeat the process 5 times. The purpose of this procedure is to have a greater concentration of cells, to ensure a greater amount of plasmid DNA to work with. | ||
| + | #Resuspend gently the bacteria in distillate water to wash away the remains of the LB medium. To resuspend means using the pipette to mix up the pellet into a solution. | ||
| + | #Pipet 250 ul of a Ribonuclease (RNase). Resuspend gently. The process of miniprep actually starts here. The enzyme RNase degrades the ARN inside the bacteria, to ensure that only DNA will be extracted. | ||
| + | #Pipet 250 ul of lytic enzyme, also called cellular lysis buffer, which will result in the lysis of the cell membrane of the bacteria. Mix by inversion 5 times. Spin with microcentrifuge for 5 minutes, at 10,000 rpm. The substance used to provoke the lysis of the bacteria varies depending on the protocol used (there are several variants of the miniprep technique). The enzyme used in this particular protocol uses a sudden and drastic change in the pH (it becomes more alkaline) of the medium of the bacteria to lysis them. The cellular contents of the bacteria are then released. Mix by inversion. | ||
| + | #Pipet 350 microliters (ul) of precipitation buffer into the solution and then mix by inversion. The precipitation buffer is used to precipitate proteins and genomic DNA. This buffer takes advantage of the fact that the DNA in the plasmid in bacteria is more supercoiled than the genomic DNA. Thus, when adding the precipitation buffer the genomic DNA and proteins inside the solution precipitate, while the plasmid DNA stays in the supernatant. The supernatant is then removed with a pipette, leaving useless substances in the pellet (proteins and genomic DNA). | ||
| + | #Spin the supernatant for 10 minutes in a microcentrifuge, at 12,000 g. | ||
| + | #Load the supernatant in the recuperation column, which has an affinity membrane that does not allow the DNA to pass through. Add the wash buffer, in which DNA is not soluble. Thus, only the wash buffer previously added will pass through the membrane, along with other substances and impurities that are not plasmid DNA (since everything left is soluble in the wash buffer, except for the desired plasmid DNA). Therefore, plasmid DNA becomes trapped in the affinity membrane. Get the liquid out with a pipette. | ||
| + | #To further ensure that the plasmid DNA sample is pure, add the TE buffer (in which DNA is very soluble). Incubate for 1 minute. Spin down with a microcentrifuge for 2 minutes, at 12,000 rpm. Since only DNA is soluble in the PE buffer, the result of this last centrifugation is that the liquid with have only plasmid DNA, and the pellet will be the impurities. | ||
| + | #Pipet out the liquid left, and head to the nanodrop to analyze the results. | ||
| + | |||
| + | |||
| + | ===='''Recuperation of Indigo'''==== | ||
| + | |||
| + | |||
| + | <ins>Summary:</ins> | ||
| + | |||
| + | Recuperating Indigo from our cells is imperative for the whole process to be worthwhile. For this the process of centrifuging can be used with an organic solvent such as chloroform or ethyl acetate. In this case we will be using chloroform. There are other methods for the recuperation of Indigo such as sonication, freezing or even can be separated using chromatography but, for practical purposes we will use the organic solvent. | ||
| + | |||
| + | |||
| + | <ins>Materials:</ins> | ||
| + | |||
| + | |||
| + | *Centrifuge that can go at 10 000 RPM | ||
| + | *Chloroform | ||
| + | *Cell cultures | ||
| + | *Test tubes | ||
| + | |||
| + | |||
| + | <ins>Protocol:</ins> | ||
| + | The first step is to take the cell cultures in their medium and centrifuge for 10 minutes at 10 000 RPM, this will separate the cells from the medium. The next step is to remove the supernatant so in the tube only cells that have already produced the indigo remain in it. Next 10 mL of chloroform are needed. The chloroform and the cell cultures must form another solution. Once this is done the tubes must go again to the centrifuge again for 10 minutes at 10 000 RPM. Once this is finished the chloroform is removed from the tube and what’s left in it is the indigo produced by the cells. In order to validate that the product is indeed indigo you can refer to the section “Detection of Indigo” included in this wiki. | ||
| + | |||
| + | |||
| + | |||
| + | ===='''Detection of the production of Indigo'''==== | ||
| + | |||
| + | |||
| + | <ins>Summary:</ins> | ||
| + | |||
| + | To confirm that the product from the enzymes introduced into the bacterial cells is indeed indigo we have decided to use the Raman Spectra. The Raman Spectra is a spectroscopic technique which is used to study vibrations, rotations and other low-frequency modes in a system. It uses monochromatic light. In this process we would measure absorbance, which is the measure of light absorbed by a suspension of bacterial cells in this case using a spectrophotometer. The spectrophotometer is used to determine the intensity of light of a wavelength transmitted by a solution, in this case our bacterial colonies. By using this we can accurately determine if we succeeded in synthesizing indigo. | ||
| + | |||
| + | |||
| + | <ins>Materials:</ins> | ||
| + | |||
| + | *Olympus 100x long working distance microscope objective. | ||
| + | *Charged-coupled device detector | ||
| + | *Holographic gratings providing a resolution of 3-5 cm-1 (1200 rulings/mm for the 785 nm line, 1800 rulings/mm for the 488 nm line). | ||
| + | *Continuous wave diode laser | ||
| + | *Spectra Physics Model 2020 BeamLock Ar+ laser | ||
| + | |||
| + | |||
| + | <ins>Protocol:</ins> | ||
| + | |||
| + | Using the Experimental Raman spectra in the dispersive mode using a Bruker Senterra Raman apparatus with two excitations, one at 785 nm and 488 nm. For this detection an Olympus 100x long working distance microscope objective will be needed as well as a charge-coupled device detector and holographic gratings providing a resolution of 3-5 cm-1 (1200 rulings/mm for the 785 nm line, 1800 rulings/mm for the 488 nm line). For the analysis at 785 nm, it is speculated that we could use a continuous wave diode laser as the excitation source and for the collection of measurements a 1 scan with an integration of 30 s could be used. For the measurements at 488 nm a Spectra Physics Model 2020 BeamLock Ar+ laser, with an output power of 0.25 mW. On this second measurements the 1 scan could have an integration time of 1000 s in order to improve the signal-to-noise ratio. | ||
| + | |||
| + | By comparing the results we obtain with the next tables we can be sure that the product we obtained from our cells is indeed indigo and this way subject it to the chemical process described above. | ||
| + | |||
| + | |||
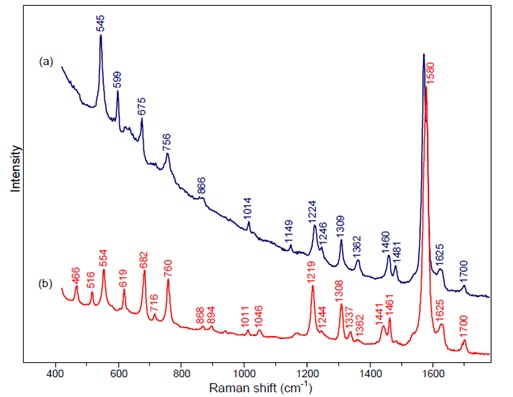
| + | [[File:figure1rec_protocols.jpg|center|Figure 1]] | ||
| + | Figure 1: this graph shows a maximum at 616 nm | ||
| + | |||
| + | |||
| + | [[File:figure2rec_protocols.jpg|center|Figure 2]] | ||
| + | Figure 2: a) indigo b) monobromoindigo comparison of Raman Spectra at 785 nm. | ||
| + | |||
| + | |||
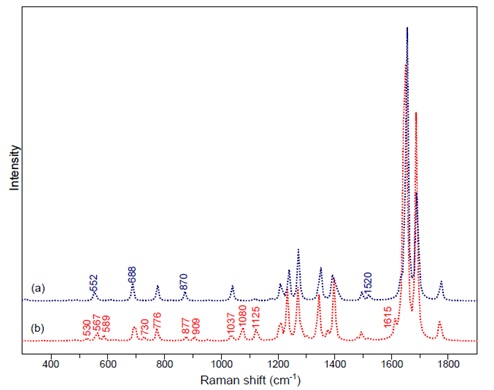
| + | [[File:figure3rec_protocols.jpg|center|Figure 3]] | ||
| + | Figure 3: DFT calculations for a) indigo b) monobromoindigo. | ||
| + | |||
| + | |||
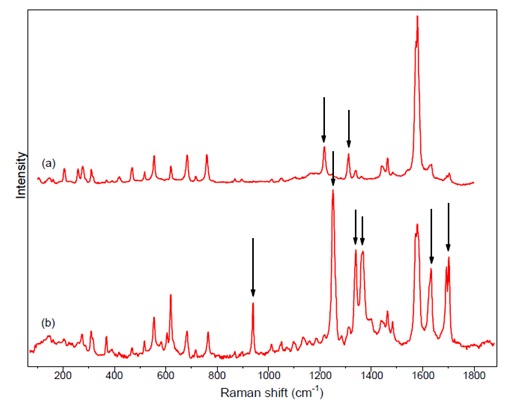
| + | [[File:figure4rec_protocols.jpg|center|Figure 4]] | ||
| + | Figure 4: comparison of experimental Raman spectra at a) 785 nm b) 488 nm, differences are highlighted by arrows | ||
| + | |||
| + | All graphs obtained from POZZI, Federica. «http://air.unimi.it/bitstream/2434/167821/2/phd_unimi_R08330.pdf.» 2012. 7 de June de 2012 <http://air.unimi.it/bitstream/2434/167821/2/phd_unimi_R08330.pdf>. | ||
| + | |||
| + | |||
| + | ===='''From indigo to Tyrian purple.'''==== | ||
| + | |||
| + | |||
| + | After obtaining the indigo, we used a chemical pathway to get the Tyrian purple. Because of the fact that from this point we only need to add some bromine atoms to the benzene rings, we will only illustrate this part of reaction with the equation below. | ||
| + | C6H6 + Br2 C6H5Br + HBr | ||
| + | FeBr3 | ||
| + | |||
| + | This reaction is called halogenation of benzene more specifically, bromination of benzene, as we are adding bromine by substitution of hydrogen. With this reaction we obtain hydrobromic acid (HBr) as a byproduct. | ||
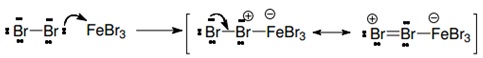
| + | The benzene ring typically does not react with halogens by itself; this is because the halogens are not enough electrophiles to interact with it. However halogens may activate themselves by means of Lewis acids. This is why we used the FeBr3, because it is considered a Lewis acid (because it can donate or share a pair of electrons), so it works as a catalyst of the reaction shown above. | ||
| + | |||
| + | The steps are as follows: | ||
| + | |||
| + | 1. Activation of the bromine by interaction with the catalyst (the Lewis acid). | ||
| + | |||
| + | [[File:step1_protocols.jpg|center]] | ||
| + | |||
| + | |||
| + | 2. The elecrophil begins to interact with the benzene ring because of the activated bromine. | ||
| + | |||
| + | [[File:step2_protocols.jpg|center]] | ||
| + | |||
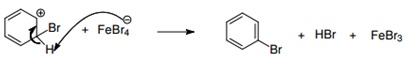
| + | |||
| + | 3. The FeBr4- acts as a base atraccting the pronton of the cation. | ||
| + | |||
| + | [[File:step3_protocols.jpg|center]] | ||
| + | |||
| + | |||
| + | It is important to state that in order for this reaction to occur, we need a low temperature and a high hydrogen pressure. | ||
</FONT COLOR> | </FONT COLOR> | ||
Latest revision as of 03:33, 17 June 2012
Contents |
Protocols
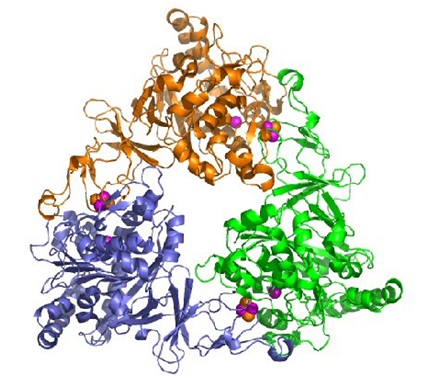
Naphthalene Dioxygenase
This enzyme is obtained from the cells of the Pseudomonas sp. which has a melting point of 55°C. One of the early uses of the enzyme was made during the process of cloning gens which were implicated in the degradation of naphthalene. In this process the fragments that were cloned of the plasmid nah express themselves while in contact with E. coli creating blue colonies. The enzyme also works as a catalyst, transforming indole into indigo, from which we obtain the tyrian purple.
What makes the indole change to indigo is an oxidation of the tryptophan that is cloned and expressed on the E. coli. The tryptophan or indole is an aromatic hydrocarbon, that when in contact with oxygen, changes to cis- dihydrodiol which makes up indigo. The oxidation is caused by combined activities of the trytophanase and napthalene diosygenase, as shown on the reaction.
The optimum ph of the naphthalene dioxygenase in the Pseudomas sp. is of 7.5, meaning that the ph is almost neutral. The optimum temperature of the enzyme is 30°C. The pressure that is exerted on the enzyme is between a reduced pressure and atmospheric pressure, which is the normal pressure we feel.
Competent cells
Summary:
As shown in the paper "Plasmid DNA purification and Agarose gel electrophoresis" written by several researchers, when you’re planning on taking a cell and make it recombinant, you need to make it competent. A competent cell is when you modified it in certain ways so that its membrane creates pores, to let DNA molecules pass through it and insert the plasmid on the cell. For this experiment, what we did was making E. Coli competent. In order to do this, first we need to prepare a Calcium chloride solution and do the next steps. The procurement of competent cells on E. Coli consists on concentrating a culture picked up on a mid-low growth logarithmic phase, and by successive washings in a cold solution (4°C) of Calcium chloride (CaCl2). Then mix the cells with the DNA. When the mixture is ready, you summit it 90 seconds to 42°C. This treating modifies the E.Coli membrane and increases its permeability to the DNA. This helps the plasmids enter the cell as a double strand molecule, while the linear DNA that enters the same way into the cell, are degraded by the Cytoplasmic nucleases (RecBCD).
Materials:
- Pipette
- LB media
- Erlenmeyer flask
- Spectrophotometer
- H2O (liquid and solid)
- Centrifuge at 4000 rpm
- CaCl2
- 50 mL tubes
Protocol Competent cells:
As shown in the paper made by Anh-Hue T. Tu , to make competent cells you should perform the following steps:
- Inoculate 5 mL of LB media with E. Coli and grow overnight at 37°C on an incubator.
- Take 1 mL of the culture growth overnight into a sterile Erlenmeyer flask with 100 mL of LB broth.
- To make sure that you’ll get most of the cells in the mid-log growth phase (5x107 cells/mL), you should take the culture into a 37°C water bath. This process should take 2 to 4 hours. The growth rate is determined by the optical density of the sample. Remove 1-ml aliquot at various times and read the optical density at 600 nm wavelength, using the spectrophotometer. The cells should be on this stage because it has been shown by several studies that in the mid-log growth phase it’s when the cells are transformed efficiently.
- When the step 3 is done, the cells are in the mid-log growth phase. Cool down the culture on ice for 10 minutes (The ice is at 4°C).
- Spin the cell suspension at 4000 rpm in a centrifuge for 5 minutes at 4°C
- Discard the supernatant.
- Resuspend the cells in half the volume of the original culture (50 mL of LB broth) with ice-cold sterile 10 mL of CaCl2.
- Place the cell suspension in an ice bath for 10 minutes.
- Centrifuge the suspension at 4000 rpm for 5 minutes at 4°C.
- Discard de supernatant.
- At the end of the suspension and resuspension, what it will be got is the pellet of cells. This pellet, at 4°C, should be next be putted on a hot water bath at 42°C. This will create pores on the membrane of the cells, allowing the DNA molecules to get into the E. Coli.
- Carefully, resuspend the cells in 10 mL of sterile ice-cold CaCl2 using precooled pipettes. The cells will remain competent for up to 24 hours at 4°C. The transformation efficiency increases four-to-six fold during this time. For long-term storage, cover a tube with aluminum, where the cells should be stored, and put in a tank with liquid nitrogen at -80°C. Depending on the strained used, the E.Coli cells will remain competent to take up DNA for as long as 6 months, making a competency test each time an aliquot is removed from storage to be used for transformation.
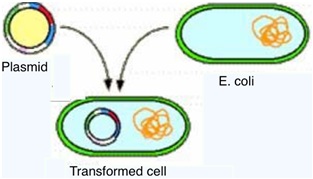
Transformation of cells
Summary:
The transformation is a process by which cells (in this case E. coli) are able to identify DNA present in the surrounding area. In order for this to take place, the E. coli has to be in a competent state.1 Some bacteria are naturally in this state, however, this is no the case of the E. coli, thus this state is achieved by following the previous process. In this state, the bacteria presents some alterations in its cellular wall and membrane, which now will allow the entrance of nucleic acids to the cell.1 The entrance of foreign DNA in the form of a plasmid –either in its circular form or in its supercoiled form- to the cell is what’s called transformation. 1 Here's an animation, made by McGraw Hill, of the process in general [http://highered.mcgraw-hill.com/sites/0072556781/student_view0/chapter14/animation_quiz_2.html].
After the cell is transformed, it has to go through the step of selection. This final step is very important, because after doing this, it’s possible that some of the bacteria didn’t take up the plasmid and therefore, in order to identify the transformed bacteria, the DNA in the plasmid has a marker.1 The marker consist in making the transformed cells resistant to the selective media. In this experiment, the media where the E. coli cells are going to be tested will contain ampicillin, and consequently only the cells with the plasmid –which will make them ampicillin resistant- are going to survive.1 By doing this, we ensure that the cells that we harvest are the ones that contain the mentioned plasmid, as they were able to develop in the selective media.1
Materials:
- Luria broth (LB)
- SOC medium
- LB plates containing ampicillin
- Filter-sterilized antibiotic solution (ampicillin)
- E. coli K12 derivatives (TB1, JM109)
- Plasmid vector: pPH219
- 1x Tris-EDTA (TE) buffer pH 8.0 (10 mM Tris Cl, 1 mM EDTA pH 8.0)
- Sterile 60 mM cold CaCl2 solution (60 mM CaCl2, 15% glycerol, 10 mM piperazine-N,N’-bis(2-hydroxypropanesulfonic acid) (PIPES), pH 7)
- Pipetters (1 to 10 μl and 10 to 200 μl range)
- Sterile pipette tips
- Sterile 50-ml glass or polypropylene tubes with caps
- 250-ml sterile Erlenmeyer flask with cap
- 5-ml glass or plastic pipettes precooled in refrigerator
- Thermometer (0 to 100°C)
- Permanent marker for labeling
- Clock or watch for timing
- 0.2-mm filter (Fisher Scientific)
- Styrofoam bucket with crushed ice
- 37°C incubator
- Water bath shaker set at 37°C
- 42°C water bath (using a thermometer ensure the temperature is exactly 42°C for the 2 minutes of heat shock)
- Centrifuge
- Roller drum in a 37°C incubator
Protocol:
- Add 10 to 40 ng (10 to 25 ml volume) of DNA to 250 ml of competent cells. Concentrated stock DNA can be diluted using sterile 1x TE buffer. This can be done to prevent a decrease in efficiency resulting from high concentration of DNA. Besides the DNA tube, additional tubes should be set up as listed below:
- In order to keep a tube denominated ¨control¨. This tube will contain 1x TE buffer in the same volume of DNA used in step 12. For the remainder of the experiment, this tube will also be submitted to the next steps.
- If competent cells were kept at -70℃, then it’s necessary to ensure that these remain able to take up DNA. The thawed cells are incubated with 10 ng of a control plasmid such as pBR322. The number of transformants per microgram of DNA will be calculated and should typically yield from 106 to 108 colonies/mg DNA for E. coli MC1061 and DH1 cells.
- Incubate the mixture on ice for 30 minutes.
- For the next 2 minutes, the mixture must be in a 42oC water bath.
- Incubate on ice for 5 minutes.
- As a next step, add 1.0 ml of SOC medium to each tube and incubate at 37oC for 1 hour in a roller drum (250 rpm). This will allow cells to express the antibiotic resistance marker, making it possible for them to survive in the selective media.
- Spread 50 to 100mL of cells on the selective media mentioned above. Store the remaining cells at 4oC.
- The E. coli cells from the control tube will be planted on selective medium and nonselective medium. In the first case, there should be no growth present, as it is a way of ensuring that, in absence of the DNA added, there would be no resistance to ampicillin, and thus the cells do not grow. On the other hand, the second plating offers viable cells that can develop in the absence of a selective medium.
- Cells that have to be tested in order to prove that they are still competent should be plated on LB agar with ampicillin (50 mg/ml final concentration). If the E. coli cells are able to survive, then it’s possible to conclude that transformation efficiency hasn’t decreased.
- Incubate all plates overnight at 37oC (agar side up).
- Count the number of colonies.
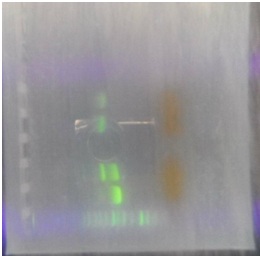
Protocol electrophoresis
Summary: In order to recover plasmids from E. Coli, you need to put it through a process known as gel electrophoresis. This technique consists on summit the DNA molecule mixture in an agarose gel with pores, and then apply an electric charge. The DNA molecules are attracted to the positive pole due to their negative charge caused by the phosphate group. The larger molecules will move slower through the pores because of their size, while the smaller molecules will move faster through the gel. When the time has gone for the molecules to separate (Around an hour and a half), the gel is submerge on an ethidium bromide solution, which has an affinity for the DNA and its intercalated in-between the base pairs and its fluorescent when its bond to it. When you illuminate the solution with UV light, you can identify the fluorescent bands which correspond to the molecules of DNA. Then, you choose the DNA molecule you need to purify and with a knife or something sharp, you cut the square where the molecule is located and take it away from the gel.
|
[http://www.phschool.com/science/biology_place/biocoach/red/gel.html Link of previous animation] |
Materials:
- Agarose
- Buffer TBE
- Sybr Green
- Microwave
- Electrophoreticbed
- Comb
- DNA ladder
- DNA simple with the desire gene
- Pipette
- UV light transiluminator
- Knife
- Power font
Protocol:
- Manufacture the agarose gel at 1%:
- On a flask, mix 1 g of agarose with 100 mL of the buffer TBE.
- Heat the mixture on a microwave till its melt and the agarose is completely dissolved.
- Add sybr green (Fluorescence) to the mixture of agarose and TBE. The fluorescence will help locate the DNA molecule of interest.
- Let the gel cool down till the temperature decreases to 50°C or 60°C. When it reaches that temperature, pour it on an electrophoretic bed. Place the comb to form the holes on the gel. On each hole, 25 μL can be pour.
- Without moving, let the mixture gellify at room temperature.
- Once the agarose gel is ready, carefully remove the comb.
- Take a DNA Ladder and mix it with a loaded dye (This dye help us see the movement of the DNA molecules when the electrophoresis has started).
- Once it has the dye, put the DNA ladder on a hole in the gel. It’s a control sample to measure the DNA molecules which have the desired length.
- Now, take the sample of DNA (25 μL) from which it will be obtained the plasmid and mixed it with a loaded dye. Put it in another one of the holes made by the comb in the gel. Repeat this process with another sample of DNA.
- Once the gel is loaded with the DNA, submit it to an electric field of 10 volts, overnight. The negative pole will be located on the side of the holes, and the positive pole on the opposite side. The molecules will move from left to right, due to the fact that DNA is charged negatively, going to the opposite pole, the positive.
- In the morning, check the electrophoresis with UV light transiluminator, locating the desired DNA molecule.
- When located cut the part of the gel where it’s located.
- Repeat steps 1 to 8, but now with an agarose gel at 3%.
Miniprep
Summary:
In order to get the plasmid DNA out of the bacteria we used the miniprep technique, which uses different solutions and a centrifuge to separate the components inside the bacterial cells. A centrifuge is essentially a device that spins contained materials extremely fast; its units are revolutions per minute (rpm). The contents inside the centrifuge become separated from one another based on their densities: denser substances will go down to the bottom of the container, while light substances and liquids go to the top. After the centrifugation, a pipette is used to remove the liquid part of the substance and thus separate it from the solid part or parts. The miniprep technique essentially takes advantage of the centrifugation technique, using solutions (usually alkaline ones) to further separate the products of the previous centrifugation. For example, the original substance is separated using centrifugation, and say, the part of interest left is in solid state. So the next step in the miniprep protocol is add a certain solution to further separate, chemically, the substance, and then to physically separate it using centrifugation. The product of this second centrifugation will be further separated, depending on its composition, by a different solution, then centrifugated again and so on, until the final or desired substance is isolated. In this case the desired substance is the bacterial plasmids, and to chemically and physically separate it is the goal of the whole miniprep technique. It is important to note that the whole process of miniprep is done using buffers or buffered solutions, because of the importance of keeping the bacteria in controlled conditions (sudden changes of pH can kill the bacteria).
Materials:
- Pipette 2 ul
- Miniprep iGEM kit
- Lysogenic Broth Medium
- Microcentrifuge
- RNase
- Lytic enzyme buffer
- Wash buffer
- PE buffer
- Recuperation columns
- Desired bacteria
Protocol:
- Pipet 1 mL of bacteria into the Lysogeny Broth medium (LB). Spin them down in a microcentrifuge for 1 minute at 10,000 rpm to separate the bacteria from the solution. Repeat the process 5 times. The purpose of this procedure is to have a greater concentration of cells, to ensure a greater amount of plasmid DNA to work with.
- Resuspend gently the bacteria in distillate water to wash away the remains of the LB medium. To resuspend means using the pipette to mix up the pellet into a solution.
- Pipet 250 ul of a Ribonuclease (RNase). Resuspend gently. The process of miniprep actually starts here. The enzyme RNase degrades the ARN inside the bacteria, to ensure that only DNA will be extracted.
- Pipet 250 ul of lytic enzyme, also called cellular lysis buffer, which will result in the lysis of the cell membrane of the bacteria. Mix by inversion 5 times. Spin with microcentrifuge for 5 minutes, at 10,000 rpm. The substance used to provoke the lysis of the bacteria varies depending on the protocol used (there are several variants of the miniprep technique). The enzyme used in this particular protocol uses a sudden and drastic change in the pH (it becomes more alkaline) of the medium of the bacteria to lysis them. The cellular contents of the bacteria are then released. Mix by inversion.
- Pipet 350 microliters (ul) of precipitation buffer into the solution and then mix by inversion. The precipitation buffer is used to precipitate proteins and genomic DNA. This buffer takes advantage of the fact that the DNA in the plasmid in bacteria is more supercoiled than the genomic DNA. Thus, when adding the precipitation buffer the genomic DNA and proteins inside the solution precipitate, while the plasmid DNA stays in the supernatant. The supernatant is then removed with a pipette, leaving useless substances in the pellet (proteins and genomic DNA).
- Spin the supernatant for 10 minutes in a microcentrifuge, at 12,000 g.
- Load the supernatant in the recuperation column, which has an affinity membrane that does not allow the DNA to pass through. Add the wash buffer, in which DNA is not soluble. Thus, only the wash buffer previously added will pass through the membrane, along with other substances and impurities that are not plasmid DNA (since everything left is soluble in the wash buffer, except for the desired plasmid DNA). Therefore, plasmid DNA becomes trapped in the affinity membrane. Get the liquid out with a pipette.
- To further ensure that the plasmid DNA sample is pure, add the TE buffer (in which DNA is very soluble). Incubate for 1 minute. Spin down with a microcentrifuge for 2 minutes, at 12,000 rpm. Since only DNA is soluble in the PE buffer, the result of this last centrifugation is that the liquid with have only plasmid DNA, and the pellet will be the impurities.
- Pipet out the liquid left, and head to the nanodrop to analyze the results.
Recuperation of Indigo
Summary:
Recuperating Indigo from our cells is imperative for the whole process to be worthwhile. For this the process of centrifuging can be used with an organic solvent such as chloroform or ethyl acetate. In this case we will be using chloroform. There are other methods for the recuperation of Indigo such as sonication, freezing or even can be separated using chromatography but, for practical purposes we will use the organic solvent.
Materials:
- Centrifuge that can go at 10 000 RPM
- Chloroform
- Cell cultures
- Test tubes
Protocol:
The first step is to take the cell cultures in their medium and centrifuge for 10 minutes at 10 000 RPM, this will separate the cells from the medium. The next step is to remove the supernatant so in the tube only cells that have already produced the indigo remain in it. Next 10 mL of chloroform are needed. The chloroform and the cell cultures must form another solution. Once this is done the tubes must go again to the centrifuge again for 10 minutes at 10 000 RPM. Once this is finished the chloroform is removed from the tube and what’s left in it is the indigo produced by the cells. In order to validate that the product is indeed indigo you can refer to the section “Detection of Indigo” included in this wiki.
Detection of the production of Indigo
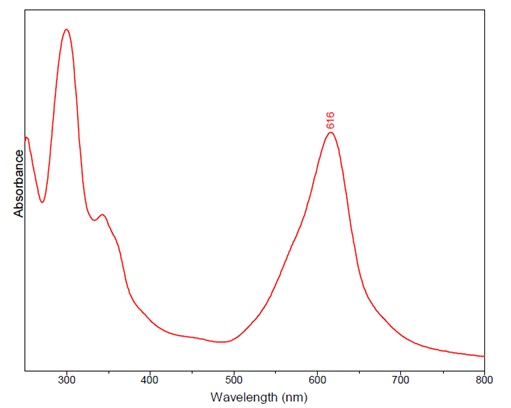
Summary:
To confirm that the product from the enzymes introduced into the bacterial cells is indeed indigo we have decided to use the Raman Spectra. The Raman Spectra is a spectroscopic technique which is used to study vibrations, rotations and other low-frequency modes in a system. It uses monochromatic light. In this process we would measure absorbance, which is the measure of light absorbed by a suspension of bacterial cells in this case using a spectrophotometer. The spectrophotometer is used to determine the intensity of light of a wavelength transmitted by a solution, in this case our bacterial colonies. By using this we can accurately determine if we succeeded in synthesizing indigo.
Materials:
- Olympus 100x long working distance microscope objective.
- Charged-coupled device detector
- Holographic gratings providing a resolution of 3-5 cm-1 (1200 rulings/mm for the 785 nm line, 1800 rulings/mm for the 488 nm line).
- Continuous wave diode laser
- Spectra Physics Model 2020 BeamLock Ar+ laser
Protocol:
Using the Experimental Raman spectra in the dispersive mode using a Bruker Senterra Raman apparatus with two excitations, one at 785 nm and 488 nm. For this detection an Olympus 100x long working distance microscope objective will be needed as well as a charge-coupled device detector and holographic gratings providing a resolution of 3-5 cm-1 (1200 rulings/mm for the 785 nm line, 1800 rulings/mm for the 488 nm line). For the analysis at 785 nm, it is speculated that we could use a continuous wave diode laser as the excitation source and for the collection of measurements a 1 scan with an integration of 30 s could be used. For the measurements at 488 nm a Spectra Physics Model 2020 BeamLock Ar+ laser, with an output power of 0.25 mW. On this second measurements the 1 scan could have an integration time of 1000 s in order to improve the signal-to-noise ratio.
By comparing the results we obtain with the next tables we can be sure that the product we obtained from our cells is indeed indigo and this way subject it to the chemical process described above.
Figure 1: this graph shows a maximum at 616 nm
Figure 2: a) indigo b) monobromoindigo comparison of Raman Spectra at 785 nm.
Figure 3: DFT calculations for a) indigo b) monobromoindigo.
Figure 4: comparison of experimental Raman spectra at a) 785 nm b) 488 nm, differences are highlighted by arrows
All graphs obtained from POZZI, Federica. «http://air.unimi.it/bitstream/2434/167821/2/phd_unimi_R08330.pdf.» 2012. 7 de June de 2012 <http://air.unimi.it/bitstream/2434/167821/2/phd_unimi_R08330.pdf>.
From indigo to Tyrian purple.
After obtaining the indigo, we used a chemical pathway to get the Tyrian purple. Because of the fact that from this point we only need to add some bromine atoms to the benzene rings, we will only illustrate this part of reaction with the equation below. C6H6 + Br2 C6H5Br + HBr
FeBr3
This reaction is called halogenation of benzene more specifically, bromination of benzene, as we are adding bromine by substitution of hydrogen. With this reaction we obtain hydrobromic acid (HBr) as a byproduct. The benzene ring typically does not react with halogens by itself; this is because the halogens are not enough electrophiles to interact with it. However halogens may activate themselves by means of Lewis acids. This is why we used the FeBr3, because it is considered a Lewis acid (because it can donate or share a pair of electrons), so it works as a catalyst of the reaction shown above.
The steps are as follows:
1. Activation of the bromine by interaction with the catalyst (the Lewis acid).
2. The elecrophil begins to interact with the benzene ring because of the activated bromine.
3. The FeBr4- acts as a base atraccting the pronton of the cation.
It is important to state that in order for this reaction to occur, we need a low temperature and a high hydrogen pressure.
 "
"