Team:Lethbridge Canada/The Project
From 2012hs.igem.org
| (13 intermediate revisions not shown) | |||
| Line 48: | Line 48: | ||
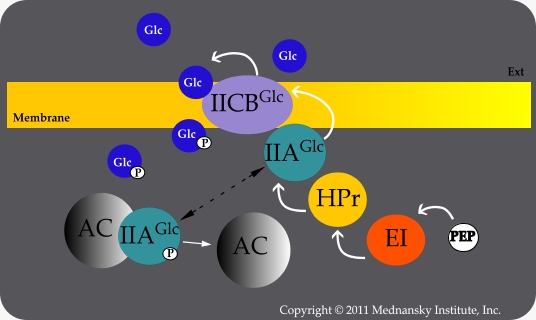
Sensory transduction is a mechanism used by many organisms to monitor their external environments. E. coli detects changes in glucose levels in its environment by means of phosphotransferase system (PTS). This system uses E. coli’s enzyme II, which consists of the membrane-bound protein EIICBGlc, as well as enzyme I (EI), a histidine phosphocarrier protein HPr, and another protein called EIIAGlc. Enzyme II is responsible for the phosphorylation and transportation of glucose into the cell. | Sensory transduction is a mechanism used by many organisms to monitor their external environments. E. coli detects changes in glucose levels in its environment by means of phosphotransferase system (PTS). This system uses E. coli’s enzyme II, which consists of the membrane-bound protein EIICBGlc, as well as enzyme I (EI), a histidine phosphocarrier protein HPr, and another protein called EIIAGlc. Enzyme II is responsible for the phosphorylation and transportation of glucose into the cell. | ||
| - | Mlc is a repressive protein that binds to a specific DNA sequence and prevents it from being transcribed. This regulates the transcription of specific genes so certain proteins are only made when they are needed. This is the mechanism we are going to use to regulate the transcription of the gene for insulin. When glucose enters the cells via enzyme II, it picks up a phosphoryl group that was originally bound to EIICBGlc. The dephosphorylated form of EIICBGlc has a high affinity with Mlc and recruits | + | Mlc is a repressive protein that binds to a specific DNA sequence and prevents it from being transcribed. This regulates the transcription of specific genes so certain proteins are only made when they are needed. This is the mechanism we are going to use to regulate the transcription of the gene for insulin. When glucose enters the cells via enzyme II, it picks up a phosphoryl group that was originally bound to EIICBGlc. The dephosphorylated form of EIICBGlc has a high affinity with Mlc and recruits Mlc from its binding site on the DNA sequence. The gene is now free to be transcribed. Due to the fact that this sequence is originally stimulated by an increase in glucose concentration as it enters the cell, it is the perfect circuit for us to use to regulate the production of inulin. |
Nam, T., Cho, S., Shin, D., Kim, J., Jeong, J., Lee, J., Roe, J., & Kang, S. (2001). The Escherichia coli glucose transport enzyme iicb(glc) recruits the global repressor mlc. | Nam, T., Cho, S., Shin, D., Kim, J., Jeong, J., Lee, J., Roe, J., & Kang, S. (2001). The Escherichia coli glucose transport enzyme iicb(glc) recruits the global repressor mlc. | ||
| Line 57: | Line 57: | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
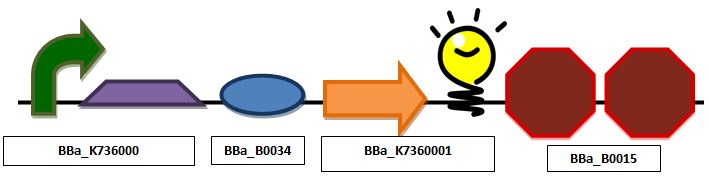
| + | ===Glucose sensing construct=== | ||
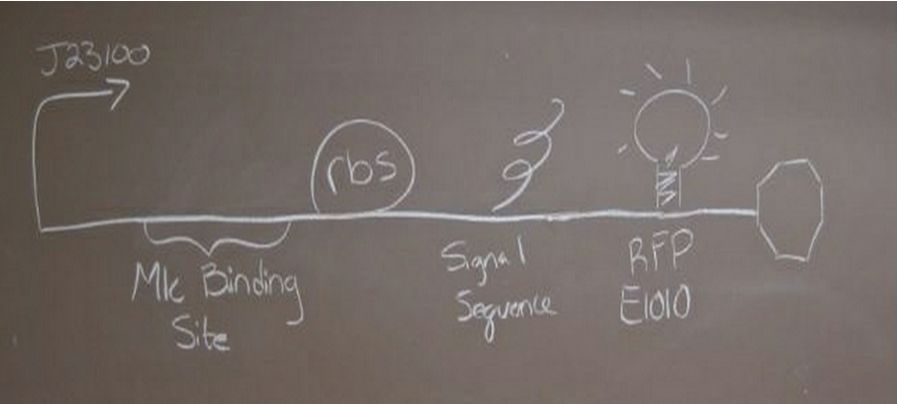
'''Construct 1 focuses on the glucose sensing and gene regulating mechanism. We will implement the global respressor Mlc in order to control the rate at which the insulin gene is transcribed. ''' | '''Construct 1 focuses on the glucose sensing and gene regulating mechanism. We will implement the global respressor Mlc in order to control the rate at which the insulin gene is transcribed. ''' | ||
[[Image:Glucose.JPG|center|px]] | [[Image:Glucose.JPG|center|px]] | ||
| Line 62: | Line 68: | ||
| - | |||
| - | |||
| + | ==Insulin Production and Secretion (Red Fluorescent Protein as Proof of Principle)== | ||
| + | This glucose sensitive promoter will be coupled to the DNA for the production of the 51 amino acid polypeptide insulin (human insulin). Insulin needs to be produced and then exported out of the cell in order for the insulin to decrease the blood glucose levels. For the export of insulin, we plan to use a signal sequence that directs the cell to transport the insulin protein outside of the cell. We will test two signal sequences: the heat stable enterotoxin I signal sequence (BBa_K331033) and the twin arginine tag (TAT) conserved motif. This can be fused to the DNA that is in charge of the insulin production. For our project, we used red fluorescent protein as a proof of principle. | ||
| - | '''Construct 2 focuses on red fluorescent production and release as a proof of principle for insulin.'''<br> | + | ===Insulin Secretion Construct=== |
| + | |||
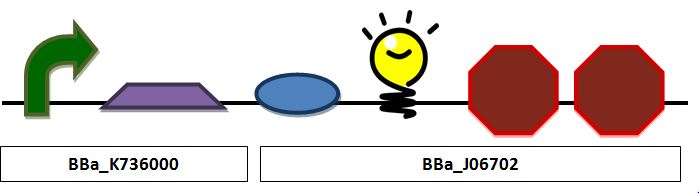
| + | '''Construct 2 focuses on red fluorescent (in place of insulin) production and release as a proof of principle for insulin.'''<br> | ||
[[Image:Insulin.JPG|center|px]] | [[Image:Insulin.JPG|center|px]] | ||
''Insulin secretion construct containing a pLac promoter, ribosomal binding site, TAT sequence fused to a red fluorescent protein, and a double terminator:'' | ''Insulin secretion construct containing a pLac promoter, ribosomal binding site, TAT sequence fused to a red fluorescent protein, and a double terminator:'' | ||
| + | |||
| + | |||
| + | |||
==Combined glucose detection and red fluorescent protein (in place of insulin) secretion== | ==Combined glucose detection and red fluorescent protein (in place of insulin) secretion== | ||
| - | + | ||
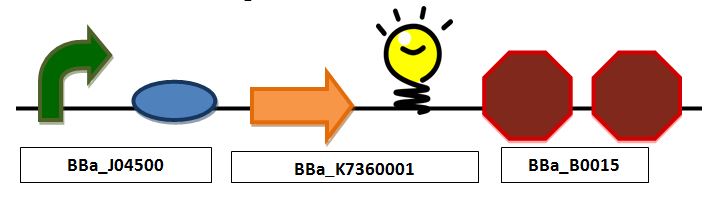
[[Image:Full.JPG|center|px]] | [[Image:Full.JPG|center|px]] | ||
| + | ''Full construct containing a promoter, mlc binding site, ribosomal bonding site, TAT signal sequence fused to a red fluorescent protein and a double terminator'' | ||
==Delivery System== | ==Delivery System== | ||
Latest revision as of 09:10, 17 June 2012

| Home | The Team | The Project | Results | Human Practices | Notebook | Safety |
|---|
Treating type I diabetes: A synthetic biology approachIntroHyperglycemic diabetes mellitus (type I diabetes) is a disorder in which pancreatic beta (insulin-producing) cells within the body are compromised, and results in the inability of the body to control glucose levels. Conventional methods of treatment can have unfavorable implications. Therefore, our team worked on using synthetic biology to derive another, potentially more viable treatment. Our project involves engineering a glucose detection and insulin production/secretion system. As a method of glucose detection, we are utilizing the natural mechanism of glucose-induced gene expression present in Escherichia coli — mlc inhibition coupled with the phosphotransferase system. The induced gene will be red fluorescent protein (RFP) as a proof of principle in place of insulin. In order to secrete insulin (or RFP respectively), we will use N-terminal signal sequences to direct targeting across the cell membrane. We are testing two—twin arginine tag and heat-stable toxin I— to determine their efficiency in secreting proteins.
DiabetesThe purpose of our project is to create bacteria that will detect blood sugar levels and respond accordingly by producing an appropriate amount of insulin. Type one diabetes is caused by the degeneration of islet cells in the pancreas. Conventional methods of treatment for type one diabetes include direct injection of insulin intravenously, the transplantation of islet cells or even the introduction of an entirely new pancreas. Our engineered bacteria would provide a long-term solution compared to the standard injections, which need to be administered at least twice per day. In essence, our project has the potential to change the way that diabetes is treated. Worldwide 346 million people suffer from diabetes, including 2.7 million people in Canada. In higher income countries diabetes usually affects people over 50 years of age the most. Lower income countries face double the mortality rate when compared to the higher income countries as a result of diabetes. Diabetes can give someone a predisposition to heart disease and 4 out of 5 diabetic patients die from heart disease. 1 in 3 people who have diabetes are unaware they have it, on average people have diabetes for 7 years before diagnosis. The cost of medication and supplies range from 1,000 to 15,000 dollars every year. It is estimated that diabetes will cost the Canadian health care system 19.2 billion dollars by 2020. (Source: Get Serious | Facts + Stats | Canadian Diabetes Association. In Canadian Diabetes Association. Retrieved June 2012, from http://www.diabetes.ca/getserious/facts/.) Glucose DetectionIt is essential that our project must include a way for Escherichia coli to detect changing glucose concentrations in its environment. If the transcription of the insulin gene is not regulated in some way then the protein will be constantly produced which will lead to an extremely high concentration of insulin in the system, which can be just as dangerous as high blood sugar levels. Sensory transduction is a mechanism used by many organisms to monitor their external environments. E. coli detects changes in glucose levels in its environment by means of phosphotransferase system (PTS). This system uses E. coli’s enzyme II, which consists of the membrane-bound protein EIICBGlc, as well as enzyme I (EI), a histidine phosphocarrier protein HPr, and another protein called EIIAGlc. Enzyme II is responsible for the phosphorylation and transportation of glucose into the cell. Mlc is a repressive protein that binds to a specific DNA sequence and prevents it from being transcribed. This regulates the transcription of specific genes so certain proteins are only made when they are needed. This is the mechanism we are going to use to regulate the transcription of the gene for insulin. When glucose enters the cells via enzyme II, it picks up a phosphoryl group that was originally bound to EIICBGlc. The dephosphorylated form of EIICBGlc has a high affinity with Mlc and recruits Mlc from its binding site on the DNA sequence. The gene is now free to be transcribed. Due to the fact that this sequence is originally stimulated by an increase in glucose concentration as it enters the cell, it is the perfect circuit for us to use to regulate the production of inulin. Nam, T., Cho, S., Shin, D., Kim, J., Jeong, J., Lee, J., Roe, J., & Kang, S. (2001). The Escherichia coli glucose transport enzyme iicb(glc) recruits the global repressor mlc.
Glucose sensing constructConstruct 1 focuses on the glucose sensing and gene regulating mechanism. We will implement the global respressor Mlc in order to control the rate at which the insulin gene is transcribed. Glucose sensing construct with a promoter, mlc binding sequence, ribosomal binding site, red fluorescent protein and double terminator
Insulin Production and Secretion (Red Fluorescent Protein as Proof of Principle)This glucose sensitive promoter will be coupled to the DNA for the production of the 51 amino acid polypeptide insulin (human insulin). Insulin needs to be produced and then exported out of the cell in order for the insulin to decrease the blood glucose levels. For the export of insulin, we plan to use a signal sequence that directs the cell to transport the insulin protein outside of the cell. We will test two signal sequences: the heat stable enterotoxin I signal sequence (BBa_K331033) and the twin arginine tag (TAT) conserved motif. This can be fused to the DNA that is in charge of the insulin production. For our project, we used red fluorescent protein as a proof of principle.
Insulin Secretion ConstructConstruct 2 focuses on red fluorescent (in place of insulin) production and release as a proof of principle for insulin. Insulin secretion construct containing a pLac promoter, ribosomal binding site, TAT sequence fused to a red fluorescent protein, and a double terminator:
Combined glucose detection and red fluorescent protein (in place of insulin) secretionFull construct containing a promoter, mlc binding site, ribosomal bonding site, TAT signal sequence fused to a red fluorescent protein and a double terminator Delivery SystemThe method of delivery of our organism can be through direct delivery in the bloodstream or through something such as the NASA Bio capsule. It is feasible to use a system that enters the blood stream because there are already millions of E. coli cells in our body, and we can use a strain that does not cause an immune response. Alternatively, the NASA Bio capsule, which is a tangle of carbon nanotubes that will be used to contain particular cells and eventually medicinal substances inside it, could also be used. The capsule could contain our cells, and, should the body need insulin, automatically start secretion. The Bio capsule is tiny, inserted into the skin, non-reactive and fast acting.
(Source: Ecach - Chapter II. In Mednansky Institute. Retrieved June 2012, from http://www.minst.org/ecoli_cyclase_Enzyme_IIA-glc.htm.) |
 "
"