Team:Dalton School NY/Notebook
From 2012hs.igem.org
(→Testing of DNA prepared by several methods) |
|||
| Line 3: | Line 3: | ||
==Testing of DNA prepared by several methods== | ==Testing of DNA prepared by several methods== | ||
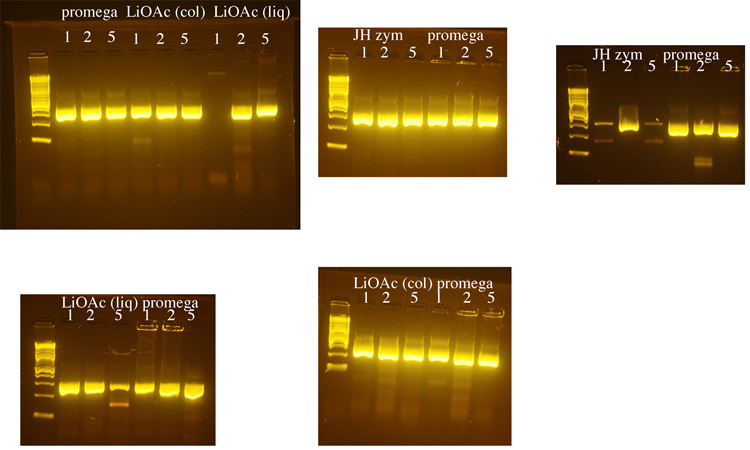
| - | Yeast genomic DNA is commonly prepared using a phenol/chloroform based method. Because both phenol and chloroform are toxic chemicals, we tested several methods for preparing yeast genomic DNA that do not use these chemicals. We compared a method that uses LiOAc to disrupt the yeast cell wall (BioTechniques 50:325-328, 2011) to methods that use the enzyme zymolyase to disrupt the cell wall (a Promega kit or a protocol from Jim Haber’s lab, “JH zym”). The Haber lab protocol produced the greatest yield of genomic DNA, but DNA made using the Promega kit was used in subsequent PCRs because it seemed to produce the most consistent results. 1µl, 2µl, or 5µl of genomic DNA was used in each PCR reaction with LIF1 promoter primers. Following these initial tests, 1µl of yeast genomic DNA was used for subsequent PCRs. The LiOAc method was the shortest and least expensive and we may switch to this method when future DNA preps are needed. | + | Yeast genomic DNA is commonly prepared using a phenol/chloroform based method. Because both phenol and chloroform are toxic chemicals, we tested several methods for preparing yeast genomic DNA that do not use these chemicals. We compared a method that uses LiOAc to disrupt the yeast cell wall (BioTechniques 50:325-328, 2011) to methods that use the enzyme zymolyase to disrupt the cell wall (a Promega kit or a protocol from Jim Haber’s lab, “JH zym”). The Haber lab protocol produced the greatest yield of genomic DNA, but DNA made using the Promega kit was used in subsequent PCRs because it seemed to produce the most consistent results. 1µl, 2µl, or 5µl of genomic DNA was used in each PCR reaction with LIF1 promoter primers. The exception was that the DNA prepared using the Haber lab protocol was diluted 1:10 because it was about 10X more concentrated than the DNA prepared using the other methods. Following these initial tests, 1µl of yeast genomic DNA was used for subsequent PCRs. The LiOAc method was the shortest and least expensive and we may switch to this method when future DNA preps for PCR are needed. |
| + | For PCR, we used: | ||
| + | |||
| + | Yeast Genomic DNA: 1µl or 2µl or 5µl | ||
| + | |||
| + | 2X OneTaq Mix (NEB): 25µl | ||
| + | |||
| + | 5µM Forward primer: 2.5µl | ||
| + | |||
| + | 5µM Reverse primer: 2.5µl | ||
| + | |||
| + | H2O: 19µl or 18µl or 15µl | ||
| + | |||
| + | |||
| + | |||
| + | '''Cycling conditions''' | ||
| + | |||
| + | 1 cycle of: | ||
| + | |||
| + | 94°C 5 min | ||
| + | |||
| + | 15 cycles of: | ||
| + | |||
| + | 94°C 30 sec | ||
| + | |||
| + | 50°C 30 sec | ||
| + | |||
| + | 68°C 45 sec | ||
| + | |||
| + | 30 cycles of: | ||
| + | |||
| + | 94°C 30 sec | ||
| + | |||
| + | 55°C 30 sec | ||
| + | |||
| + | 68°C 45 sec | ||
| + | |||
| + | 1 cycle of: | ||
| + | |||
| + | 68°C 5 min | ||
[[Image:Dalton_Fig7.jpg]] | [[Image:Dalton_Fig7.jpg]] | ||
| Line 24: | Line 63: | ||
H2O: 19µl | H2O: 19µl | ||
| + | |||
| + | |||
| + | |||
| + | '''Cycling conditions''' | ||
| + | |||
| + | 1 cycle of: | ||
| + | |||
| + | 94°C 5 min | ||
| + | |||
| + | 15 cycles of: | ||
| + | |||
| + | 94°C 30 sec | ||
| + | |||
| + | 50°C 30 sec | ||
| + | |||
| + | 68°C 45 sec | ||
| + | |||
| + | 30 cycles of: | ||
| + | |||
| + | 94°C 30 sec | ||
| + | |||
| + | 55°C 30 sec | ||
| + | |||
| + | 68°C 45 sec | ||
| + | |||
| + | 1 cycle of: | ||
| + | |||
| + | 68°C 5 min | ||
| Line 275: | Line 342: | ||
H2O: 18µl | H2O: 18µl | ||
| + | |||
| + | |||
| + | '''Cycling conditions''' | ||
| + | |||
| + | 1 cycle of: | ||
| + | |||
| + | 94°C 5 min | ||
| + | |||
| + | 15 cycles of: | ||
| + | |||
| + | 94°C 30 sec | ||
| + | |||
| + | 50°C 30 sec | ||
| + | |||
| + | 68°C 45 sec | ||
| + | |||
| + | 30 cycles of: | ||
| + | |||
| + | 94°C 30 sec | ||
| + | |||
| + | 55°C 30 sec | ||
| + | |||
| + | 68°C 45 sec | ||
| + | |||
| + | 1 cycle of: | ||
| + | |||
| + | 68°C 5 min | ||
Revision as of 20:08, 1 June 2012
In progress...
Contents |
Testing of DNA prepared by several methods
Yeast genomic DNA is commonly prepared using a phenol/chloroform based method. Because both phenol and chloroform are toxic chemicals, we tested several methods for preparing yeast genomic DNA that do not use these chemicals. We compared a method that uses LiOAc to disrupt the yeast cell wall (BioTechniques 50:325-328, 2011) to methods that use the enzyme zymolyase to disrupt the cell wall (a Promega kit or a protocol from Jim Haber’s lab, “JH zym”). The Haber lab protocol produced the greatest yield of genomic DNA, but DNA made using the Promega kit was used in subsequent PCRs because it seemed to produce the most consistent results. 1µl, 2µl, or 5µl of genomic DNA was used in each PCR reaction with LIF1 promoter primers. The exception was that the DNA prepared using the Haber lab protocol was diluted 1:10 because it was about 10X more concentrated than the DNA prepared using the other methods. Following these initial tests, 1µl of yeast genomic DNA was used for subsequent PCRs. The LiOAc method was the shortest and least expensive and we may switch to this method when future DNA preps for PCR are needed.
For PCR, we used:
Yeast Genomic DNA: 1µl or 2µl or 5µl
2X OneTaq Mix (NEB): 25µl
5µM Forward primer: 2.5µl
5µM Reverse primer: 2.5µl
H2O: 19µl or 18µl or 15µl
Cycling conditions
1 cycle of:
94°C 5 min
15 cycles of:
94°C 30 sec
50°C 30 sec
68°C 45 sec
30 cycles of:
94°C 30 sec
55°C 30 sec
68°C 45 sec
1 cycle of:
68°C 5 min
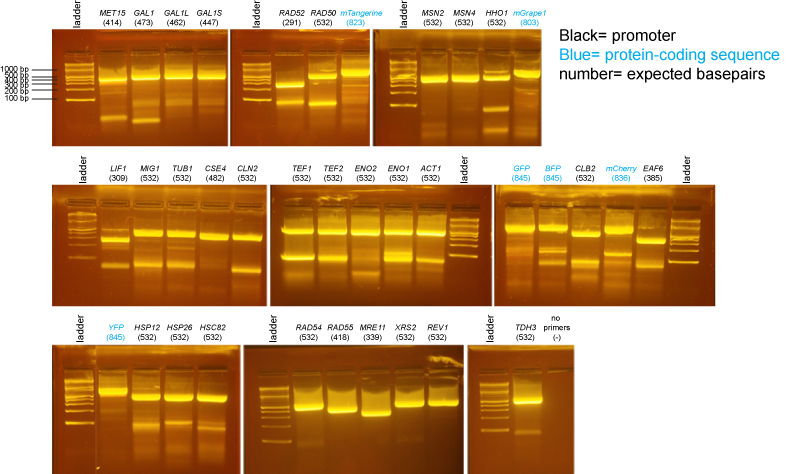
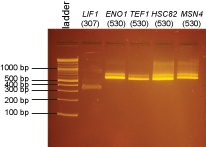
PCR of promoters and fluorescent protein coding sequences
We did an isolation of genomic DNA from yeast using the Promega Wizard Genomic DNA Purification Kit- cat# A1120. We substituted 20T zymolyase (Seikagaku) for lyticase.
For PCR, we used:
Yeast Genomic DNA: 1µl
2X OneTaq Mix (NEB): 25µl
5µM Forward primer: 2.5µl
5µM Reverse primer: 2.5µl
H2O: 19µl
Cycling conditions
1 cycle of:
94°C 5 min
15 cycles of:
94°C 30 sec
50°C 30 sec
68°C 45 sec
30 cycles of:
94°C 30 sec
55°C 30 sec
68°C 45 sec
1 cycle of:
68°C 5 min
The ladder on all gels is the 2-log ladder (NEB).
2% agarose gels in 1X Sodium Borate buffer: BioTechniques 36:214-216 (February 2004)
There may be more nonspecific bands in these PCR reactions than is typical either because there was a considerable delay waiting for all students to be ready to add their tubes to the PCR machine or because we had an issue where the program was initially incorrect and we needed to restart the PCR machine after correcting the problem.
Then we conducted a gel extraction, using QIAquick Gel Extraction Kit.
Blunting the PCR Product
After we had the purified PCR product, we used the Blunt Cloning Kit (NEB) to blunt and phosphorylate the ends of the PCR product. The blunting reaction was composed of:
Purified DNA: 15µl
10X Blunting Buffer: 2.5µl
1 mM dNTP Mix: 2.5µl
Blunt Enzyme Mix: 1.0µl
Sterile dH20: 4µl
Total Volume: 25µl
Digestion of pUC19
The pUC19 vector was digested with SmaI:
1µg/µl pUC19 (NEB): 5µl
10X NEBuffer 4: 3µl
SmaI: 1 µl
H2O: 11 µl
The restriction digest was incubated for 4 hours at 25°C. 1µl of Calf Intestinal Phosphatase (NEB) was added and the mixture was incubated at 37°C for 2 hrs.
Ligation
Next, we did a ligation reaction with the Quick Ligation Kit (NEB). We followed the manufacturer's suggested protocol which was to use a 3:1 molar ratio of insert: vector and 50 ng of SmaI digested/CIP'd pUC19 vector.
Transformation
For the transformation, we used NEB 10-beta Competent E. coli cells (cat # C3019H). We followed the manufacturer's suggested protocol for the transformation, and we plated on Ampicillin/IPTG/X-Gal.
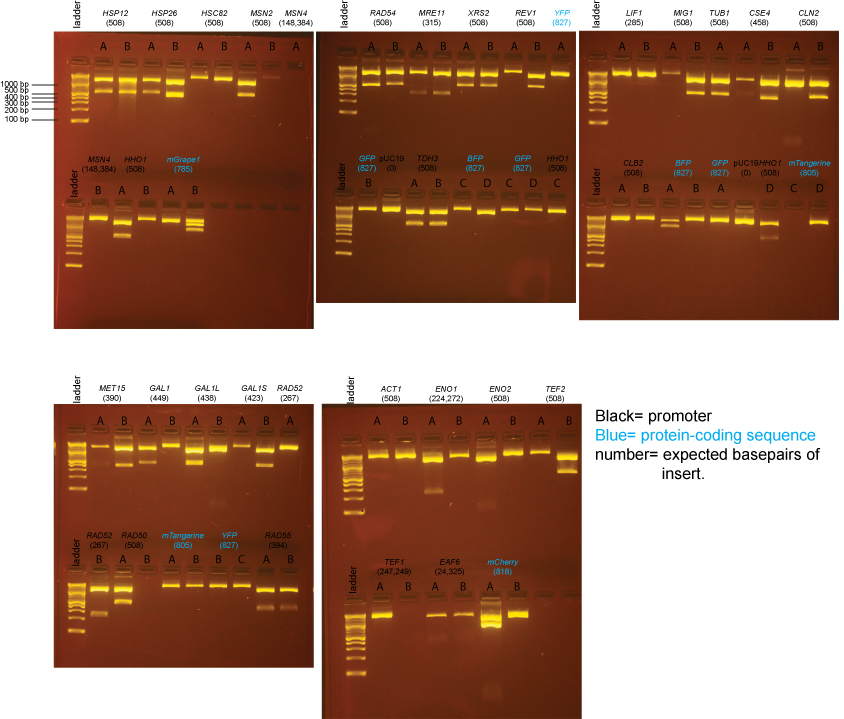
BsaI-digested Miniprep DNA
Overall, most plates contained a mixture of blue and white colonies and the number of colonies was fairly low - not much higher than for our "no insert" ligations. We then picked two white colonies from each plate and inoculated 2ml of LB/amp and then mini-prepped the DNA. Blue colonies had intact lacZ genes, and therefore did not contain inserts. The samples on these gels contain BsaI-digested miniprep DNA prepared from individual colonies from the transformations above.
For the BsaI Digest, we used:
Miniprep DNA: 4µl
10X NEB4: 2µl
100X BSA: .2µl
BsaI: .5µl
H20: 13.3µl
Expected band sizes from BsaI digests
pUC19 (no insert): 1 cut, 1 band: 2686 bp
pUC19 + insert: 3 cuts, 3 bands: 1354 bp, 1332 bp, insert (expected sizes listed on gel below)
Note that the 1354bp and 1332bp bands will appear to run as one band on the gel.
Based on the data above, we concluded that the following clones had inserts of the correct size:
Promoters
MET15 yes: B
GAL1 yes: A
GAL1-L yes: A
GAL1-S yes: B
RAD52 yes: B
RAD50 yes: A
RAD54 yes: A, B
RAD55 yes: A, B
MRE11 yes: A, B
XRS2 yes: A, B
REV1 yes: B
TDH3 yes: A, B
LIF1 none
MIG1 yes: B
TUB1 yes: A
CSE4 yes: B
CLN2 yes: B
CLB2 none
ACT1 none
ENO1 none
ENO2 none
TEF2 yes: B
TEF1 none
EAF6 none (possibly a band, but too little DNA to be confident)
HSP12 yes: A, B
HSP26 yes: A
HSC82 none
MSN2 yes: A
MSN4 none
HHO1 yes: A, D
Protein-coding sequences
BFP maybe: A (the insert may be too large)
GFP none
YFP none
mTangerine none
mCherry yes: A (but bottom band seems possibly too intense)
mGrape1 none
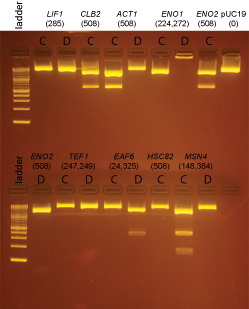
Extra colonies were picked from the transformations for the following promoter clones:
LIF1, CLB2, ACT1, ENO1, ENO2, TEF1, EAF6, HSC82, MSN4
DNA from these clones was miniprepped using Qiagen minipreps. The DNA was digested with BsaI as follows:
miniprep DNA: 3µl
10X NEBuffer 4: 2µl
100X BSA: 0.2µl
BsaI: 0.5µl
H2O: 14.8µl
BsaI digests were incubated at 37°C for 2 hrs and then run on a 2% agarose gel in 1X sodium borate buffer.
Based on the data above, we concluded that the following clones had inserts of the correct size:
Promoters
LIF1 none
CLB2 yes: C
ACT1 yes: C
ENO1 none
ENO2 yes: C
TEF1 none
EAF6 yes: D (This was started from culture EAF6-B)
HSC82 none
MSN4 none
Obtaining the remaining clones
Because there had been relatively few colonies on most of the transformation plates, we decided to try 2 different strategies for obtaining the remaining clones:
1. Repeat the original blunt ligations using a higher insert:vector ratio. 2. Try a sticky-end ligation strategy since it may have a higher success rate than the blunt ligation strategy. (This strategy is outlined in detail below:
When we designed the PCR primers, we added nucleotides 5' to the BSA recognition site in both the forward and reverse primers. We decided to introduce the sequence 5' CCCGGG 3'which is the site for both SmaI (creates blunt ends) and XmaI (creates sticky ends). According to the NEB catalog, SmaI and XmaI can cut very close to the end of a PCR product. Thus, the introduction served as a backup strategy if the blunt cloning was problematic. The PCR products could be cut with XmaI and ligated into XmaI-digested pUC19.
The original PCR reaction was digested with XmaI for the following promoters:
LIF1, ENO1, TEF1, HSC82, MSN4
purified PCR product: 15µl
10X NEBuffer 4: 3µl
100X BSA: 0.3µl
XmaI: 0.5µl
H2O: 11.2µl
These digested PCR products were run on the gel below:
2% agarose gel in 1X sodium borate buffer:
These bands were excised and purified using the QiaQuick Gel Extraction Kit.
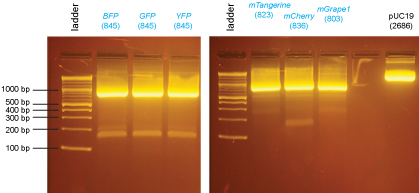
Because the cloning of the fluorescent proteins was particularly problematic, we repeated the PCR of these genes:
5ng/µl fluorescent protein vector: 2µl
2X OneTaq Mix (NEB): 25µl
5µM Forward primer: 2.5µl
5µM Reverse primer: 2.5µl
H2O: 18µl
Cycling conditions
1 cycle of:
94°C 5 min
15 cycles of:
94°C 30 sec
50°C 30 sec
68°C 45 sec
30 cycles of:
94°C 30 sec
55°C 30 sec
68°C 45 sec
1 cycle of:
68°C 5 min
PCR products were purified using the QiaQuick Gel Extraction Kit.
The purified PCR products were then digested with XmaI:
purified PCR product: 15µl
10X NEBuffer 4: 3µl
100X BSA: 0.3µl
XmaI: 0.5µl
H2O: 11.2µl
These digested PCR products were run on the gel below and the bands were excised and purified using the QiaQuick Gel Extraction Kit.
The pUC19 vector was digested with XmaI:
1µg/µl pUC19 (NEB): 5µl
10X NEBuffer 4: 3µl
100X BSA: 0.3µl
XmaI: 0.5µl
H2O: 11.2µl
The pUC19 vector was digested overnight at 37°C. Then, the reaction was heated at 65°C for 20min. 1µl of Calf Intestinal Phosphatase (NEB) was added to the reaction and it was incubated at 37°C for 2 hrs. This digested, dephosphorylated vector was run on the agarose gel below.
2% agarose gel in 1X sodium borate buffer:
These bands were excised and purified using the QiaQuick Gel Extraction Kit.
Strategy 1: blunted PCR products were ligated into SmaI/CIP'd pUC19 vector as before, except that a 10:1 molar insert:vector ratio was used.
Strategy 2: XmaI-digested PCR products were ligated into XmaI/CIP'd pUC19 vector using a 6:1 molar insert:vector ratio.
5µl of ligation reaction were transformed and plated on LB/Amp/X-gal/IPTG.
Return to our main page.
 "
"