Team:Dalton School NY/Notebook
From 2012hs.igem.org
| Line 10: | Line 10: | ||
[[Image:Dalton_Fig6.jpg]] | [[Image:Dalton_Fig6.jpg]] | ||
| + | |||
| + | ==BsaI-digested Miniprep DNA == | ||
| + | |||
| + | The samples on these gels contain BsaI-digested miniprep DNA prepared from individual colonies from the transformations above. | ||
| + | |||
| + | [[Image:Dalton_Fig8.jpg]] | ||
Return to our [https://2012hs.igem.org/Team:Dalton_School_NY main page]. | Return to our [https://2012hs.igem.org/Team:Dalton_School_NY main page]. | ||
Revision as of 02:26, 1 June 2012
In progress...
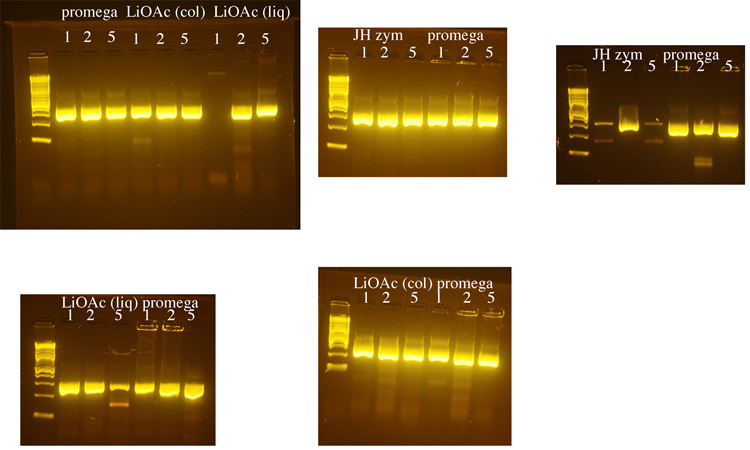
Testing of DNA prepared by several methods
Yeast genomic DNA is commonly prepared using a phenol/chloroform based method. Because both phenol and chloroform are toxic chemicals, we tested several methods for preparing yeast genomic DNA that do not use these chemicals. We compared a method that uses LiOAc to disrupt the yeast cell wall (BioTechniques 50:325-328, 2011) to methods that use the enzyme zymolyase to disrupt the cell wall (a Promega kit or a protocol from Jim Haber’s lab, “JH zym”). The Haber lab protocol produced the greatest yield of genomic DNA, but DNA made using the Promega kit was used in subsequent PCRs because it seemed to produce the most consistent results. 1µl, 2µl, or 5µl of genomic DNA was used in each PCR reaction with LIF1 promoter primers. Following these initial tests, 1µl of yeast genomic DNA was used for subsequent PCRs. The LiOAc method was the shortest and least expensive and we may switch to this method when future DNA preps are needed.
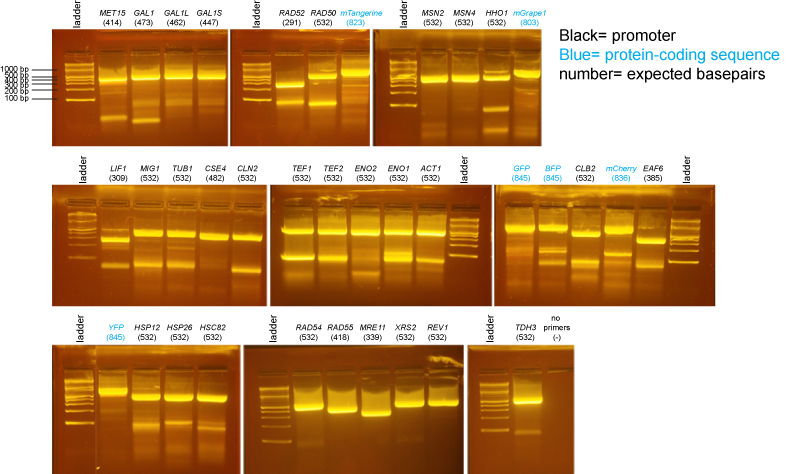
PCR of promoters and fluorescent protein coding sequences
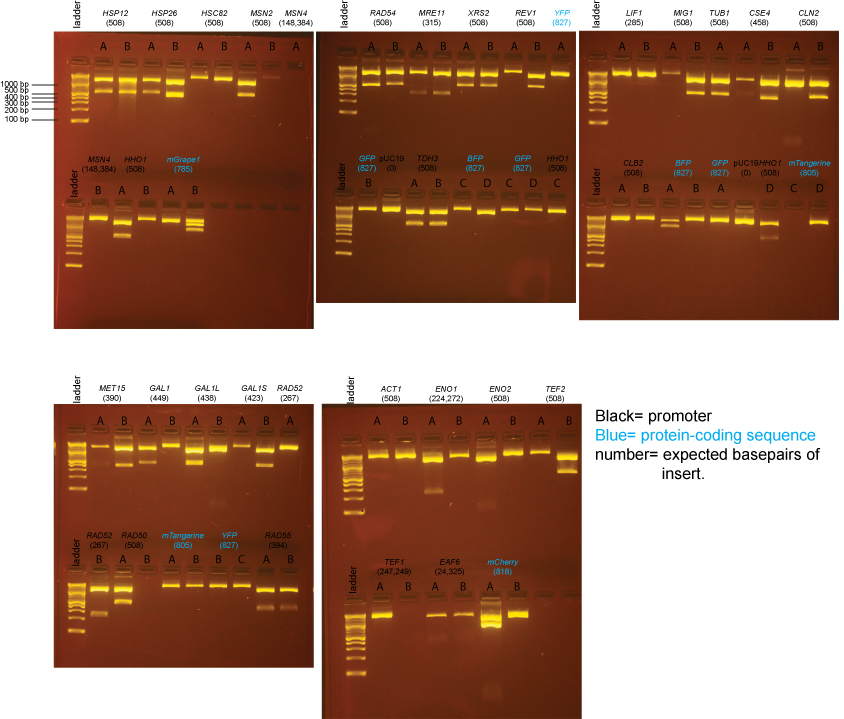
BsaI-digested Miniprep DNA
The samples on these gels contain BsaI-digested miniprep DNA prepared from individual colonies from the transformations above.
Return to our main page.
 "
"