Team:CSIA SouthKorea/results
From 2012hs.igem.org
(Difference between revisions)
Eugeniestar (Talk | contribs) (→Results & Conclusion) |
(→Experiment Results) |
||
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
==Simulation Results== | ==Simulation Results== | ||
| + | |||
| + | : Before doing the wet lab, our team did [[Team:CSIA_SouthKorea/simulation | simulation]]. However, the results of our simulation turned out to be somewhat unexpected. Therefore, we tried to find the problem in our simulation code. | ||
| + | |||
| + | |||
| + | :: public static boolean finish (Double[] in, int[] areas, Double err, int count){ | ||
| + | ::: Double [] imp = new Double[count]; | ||
| + | ::: for (int i=0; i<count; i++){ | ||
| + | |||
| + | :::: imp[i] = in[areas[i]]; | ||
| + | |||
| + | ::: } | ||
| + | ::: if (equal(imp,err,count)) | ||
| + | :::: return false; | ||
| + | ::: return true; | ||
| + | :: } | ||
| + | |||
| + | |||
| + | : This part of the code indicates to print the time when the values of the function of selected cells are equal to each other. However, what we wanted to do was printing the time when the values of the function of selected cells are all similar to 1, inside the acceptable error bound. Some parts of the code that misleads the result of the simulation, like the part indicated above, should be debugged for improvement, which we are planning to complete soon. | ||
| + | |||
| + | : Furthermore, in order to simplify the problem, we simplified the oscillation of GFP expression into a sine function; however, actually, the oscillation does not follow such simple function, but rather follows a more complex differential equation function. To represent the problem more accurately, we will try to fix those parts as well soon. | ||
==Experiment Results== | ==Experiment Results== | ||
| + | |||
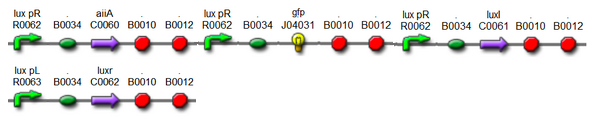
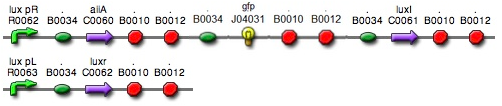
| + | : Due to complexity in our model, we had to change our assembly methods for several times. Our initial model contained three luxpR promoters, unlike current one which has only one luxpR promotor. Since linking all of the biobrick parts was a bulky process, after confirming that RBS-aiia-RBS-GFP-RBS-luxI sequence was short enough to be expressed under single luxpR promoter, we decided to go with single luxpR promotor as in a current model. | ||
| + | |||
| + | original model | ||
| + | : [[File:original.jpg]] | ||
| + | |||
| + | modified model | ||
| + | : [[File:modified.jpg]] | ||
| + | |||
| + | : Further, we also modified some other assembly methods. We first went with standard assembly in connecting each of the biobrick parts, but due to the short RBS sequence, we had to change that part to gibson assembly. So we used Gibson assembly in constructing these parts shown below. | ||
| + | |||
| + | : [[File:pink.jpg]] | ||
| + | |||
| + | : However, there was no problem in using 3A assembly methods in further steps. Our cloning was successful, but we did not yet cultured cells and observed their oscillation periods. | ||
| + | |||
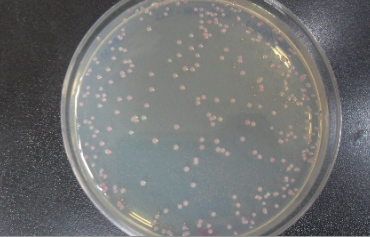
| + | : This is the result of cloning, which we had a few white colonies as we expected. | ||
| + | : [[File:colony.jpg]] | ||
| + | |||
| + | : Until the jamboree, we're planning to culture cells in array and compare the result to our simulations and expectations. | ||
Latest revision as of 03:57, 17 June 2012
Simulation Results
- Before doing the wet lab, our team did simulation. However, the results of our simulation turned out to be somewhat unexpected. Therefore, we tried to find the problem in our simulation code.
- public static boolean finish (Double[] in, int[] areas, Double err, int count){
- Double [] imp = new Double[count];
- for (int i=0; i<count; i++){
- public static boolean finish (Double[] in, int[] areas, Double err, int count){
- imp[i] = in[areas[i]];
- }
- if (equal(imp,err,count))
- return false;
- return true;
- }
- This part of the code indicates to print the time when the values of the function of selected cells are equal to each other. However, what we wanted to do was printing the time when the values of the function of selected cells are all similar to 1, inside the acceptable error bound. Some parts of the code that misleads the result of the simulation, like the part indicated above, should be debugged for improvement, which we are planning to complete soon.
- Furthermore, in order to simplify the problem, we simplified the oscillation of GFP expression into a sine function; however, actually, the oscillation does not follow such simple function, but rather follows a more complex differential equation function. To represent the problem more accurately, we will try to fix those parts as well soon.
Experiment Results
- Due to complexity in our model, we had to change our assembly methods for several times. Our initial model contained three luxpR promoters, unlike current one which has only one luxpR promotor. Since linking all of the biobrick parts was a bulky process, after confirming that RBS-aiia-RBS-GFP-RBS-luxI sequence was short enough to be expressed under single luxpR promoter, we decided to go with single luxpR promotor as in a current model.
original model
modified model
- Further, we also modified some other assembly methods. We first went with standard assembly in connecting each of the biobrick parts, but due to the short RBS sequence, we had to change that part to gibson assembly. So we used Gibson assembly in constructing these parts shown below.
- However, there was no problem in using 3A assembly methods in further steps. Our cloning was successful, but we did not yet cultured cells and observed their oscillation periods.
- Until the jamboree, we're planning to culture cells in array and compare the result to our simulations and expectations.
 "
"