Team:CSIA SouthKorea
From 2012hs.igem.org
Our team is consisted of four students who are fond of thinking creatively, sharing our knowledge with others and making contributions to the society. We hope that iGEM 2012 could be a great opportunity for us to get our feet wet in the field of synthetic biology and interact with many other students around the world who are also interested in this field!
Contents |
Team
Project
Abstract
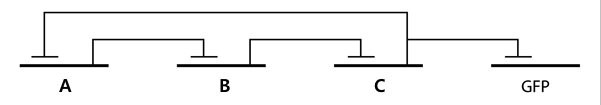
- Based on the design of V.fischeri, we placed luxR gene under luxpL promotor and placed luxI, Aiia, and GFP gene under luxpR promotor. In this V.fischeri quorum sensing system, LuxI synthase produces an acyl-homoserine lactone (AHL), which is a small molecule diffuses extracellularly and triggers quorum sensing. When AHL binds to LuxR, it produces LuxR–AHL complex that activates luxI promoter1. This also activate GFP genes, so fluorescence can be detected. AiiA 'represses' continuing activation of luxI promotor by degrading of AHL. Therefore, fluorescence may have the cycle under right conditions.
- In the world where people suffer from energy deficiency, we expect that this technology could be applied to many different areas. Among them, we think the most successful adaptation would be as an alternative for light bulbs such as those in night stand.
- We got interested in synchronized oscillator while reading Team Wageningen's 2011 project. However, we modified their model a little to increase the probability of success in experiment by using only one promoter.
Introduction of E.Coli display using repressilator
- The main mechanism that we are planning to use is the repressilator. Repressilator is a synthetic genetic regulatory network. It was reported by Michael Elowitz and Stanislas Leibler in 2000. This network is composed of 3 genes connected in a feedback loop, and each of the genes is repressed by the previous gene. This system is designed to exhibit a stable oscillation in the expression of each gene with fixed time period. In each of the wells in the 96 well plate, a colony of Escherichia Coli that expresses fluorescent protein with repressilator system will be put in. Then, by putting in different inducers in each of the wells, we are planning to control the time period and the gene expression of the E. Coli colonies and therefore express the specific shape that we are trying to express withe E. Coli display.
- Using this mechanism of repressilator to make time difference in expressing GFP, we would be able to make a certain figure on a 96 well plate. However, the periods of GFP expression in each cell will be slightly different, so we thought of applying quorum sensing to synchronize the periods of bacterias.
- picture of a 96 well plate (http://avena.pw.usda.gov)
- Our project has several steps, including cloning the repressilator, determining the variable that controls the period of oscillation, and cultivating E.Coli in a batch culture.
Mechanism of the circuit
- Our construction of genetic circuit is based on "A synthetic oscillatory network of transcriptional regulators" by Michael B. Elowitz & Stanislas Leibler, and the article “A synchronized quorum of genetic clocks” by Danino et al.
- In the diagram above, luxpL is a constitutive promoter and continuously expresses luxR.
- luxpR is an inducible promotor that is activated by luxR-AHL complex. AHL is the auto-inducer molecule N-(3-oxohexanoyl)-homoserine lactone that can diffuse across other cell membranes, gives many cells identical AHL levels, and creates quorum sensing by synchronizing AHL dependent gene expression. Under luxpR, there are both luxI, which produces AHL, and aiia, which degrades AHL, which generates feedback loops that controls the amount of luxR-AHL complex. (LuxI only increases level of AHL, which makes positive feedback loop, and aiia degenerates AHL, which builds negative feedback loop)
- Green Fluorescent protein is also under luxpR and demonstrates AHL concentration through its brightness at certain time point. Because rate of synthesis in luxI and rate of degradation in aiia differs, there exists a certain condition that enables periodic oscillation in AHL concentration.
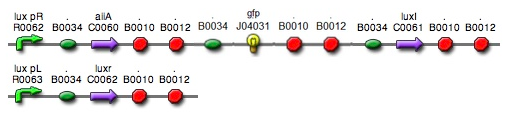
- The parts we used in the assembly are as follows: BBa_R0062 for luxpR, BBa_R0063 for luxpL, BBa_C0060 for aiia, BBa_C0061 for luxI, and BBa_K082006 for luxR. We changed the part for luxR because we found that this part, although previously used by Wageningen_UR in 2011, is inconsistent and is not confirmed.
Design of the circuit
- Since gene parts were already on the partsregistry, we linked them using Gibson assembly. However, our model is different from original plasmids used by Danino et al. We used luxpR only once to bring aiia, GFP, and luxI protein under the promoter. And we used luxpL promoter only once, placing luxR under it.
Variables that determine period of the circuit
- According to the thesis 'A synthetic oscillatory network of transcriptional regulators' by Michael B. Elowitz & Stanislas Leibler, he period of oscillations in such networks is determined mainly by the stability of the protein that is expressed by the synchronized oscillators.
- Further, 'A synchronized quorum of genetic clocks' by Danino et al. suggests that effective AHL dissipation rate affects the period of the oscillations. In other words, this means that flow rate significantly affects period. According to their experiment, at high flow rate, the stabilized oscillations exhibit period of 90+-6 min and mean amplitude of 54+-6 GFP arbitrary units. At low flow rate, they observed a period of 55 +-6min and amplitude of 30 +-9GFP arbitrary units. Overall, when they changed the flow rate from 180 to 296 micrometer per minute, the team observed an increasing oscillatory period from 52 to 90 min.
- Both factors are beyond our control as we do not have proper equipments to control both factors.
Simulation of the display
- Before doing the wet lab, in order to predict the results of our experiment, we did computer simulation of the light bulbs of the night stand that we are trying to make with colonies of Escherichia coli.
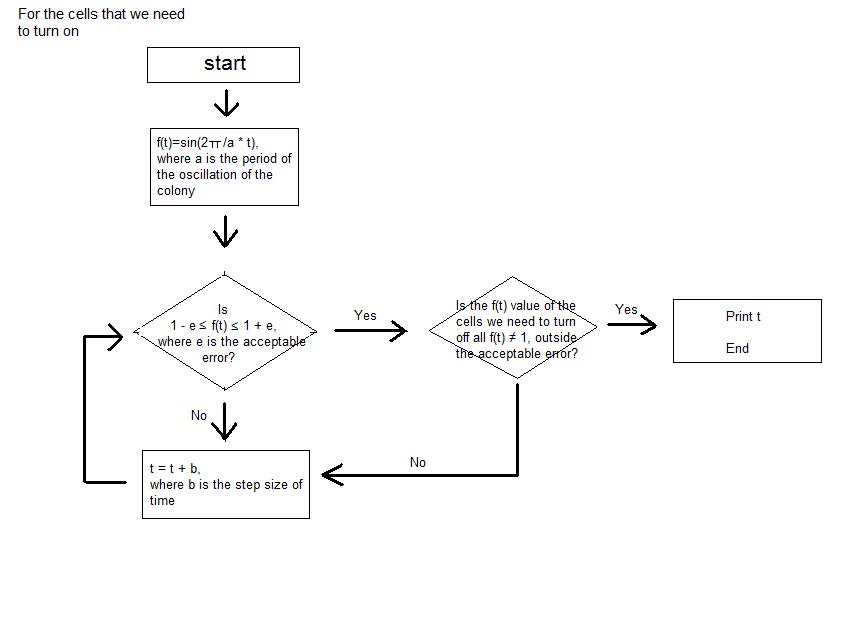
- We simplified the oscillations of GFP expression in each E. coli colonies into sine function on time. Therefore, when the periods of the oscillation of the E. coli colonies are entered as input, in each cell of the 3*3 array, which represents the 9-well-plate, the sine function with each of the entered period is corresponded. We set -1 as the value of the sine function when the GFP expression is the lowest and 1 as the value of the sine function when the GFP expression is the highest. We assumed that the GFP expression of all the colonies started from the sine function value of 0.
- Then, when the step size of time, tolerance of error, the picture that we are trying to express with the night stand are entered as the input, the program calculates the sine function value of each cell at certain time and seeks for the point where only the colonies of certain cells have the sine function value close to 1 inside the tolerance of error that was entered as input. Until the point is reached, the program keeps on calculating the sine function value of each colony by increasing the elapsed time by the step size of time that was entered as input.
- The program continuously shows the process; we can actually see the numerical value of sine function of each cell at each moment. The program stops running at the time when the picture that we want to obtain (when the certain cells that make the picture have sine function values very close to 1) is expressed, and the program prints the elapsed time as output.
- As a result, we are able to calculate the time needed to express the picture that we want to obtain using the light bulbs (colonies of E. coli in each cell) of the night stand (9-well-plate).
- This is the instruction of how the values are put into the program.
Protocols
- Our protocols have changed several times. First, we have been strictly following standard assembly, according to Biobrick Assembly Manual from ginkgobioworks and New England Biolabs.
Applications
- 1. Electronic display
- While working on the project, we wondered if we could synchronize the period of each cell in the well plate and let them have the same period of oscillation. If this is possible, then we would be able to let the bacteria glow and turn off the light in the same period, and therefore use it to make certain signals that we want on the well plate. It can be some shapes such as square, circle, cross, or star; it can be some letters such as a, b, or c; and if the technique can be applied to larger plate, then we would even be able to display a phrase or a short sentence on the plate. This might as well work as electronic displays that are widely used nowadays for advertising the stores.
- If the glowing bacteria-project that we are working on can be produced with enough light intensity and possible to program the cells of the plate, then it will be truly beneficial both for people and the environment. Currently the electricity deficiency is a huge problem all over the world. Great dark-outs occur because of excessive usage of electricity, and people are paying a great amount of money for using the electricity. If we can replace these electronic displays with glowing bacteria-display, then we will be able to save quite amount of electricity usage and thus contribute to the saving of electricity.
- 2. Light source
- Second possible application is the usage of glowing bacteria as the light sources, such as reading lamps or small-sized flashlights. It is true that certain level of light intensity is necessary in order to be used in the way proposed. However, we do believe that after some extensive research and laboratory, enough level of light intensity will be reached.
- If the light source using biological method (glowing bacteria) is developed, it will have positive impact on the society. First, since it glows with no need of external power source, battery and electricity will be saved for other usages. Second, the biological-light source will be able to be used by people in the regions where electricity supply isn’t enough or easy. Even now, there are regions where people can’t easily get access to electricity, and thus have to live in dark when the sun sets. If our project can be developed and applied to making a self-glowing light source, then it will surely bring some benefits.
Outreach & contribution
- We think it is important for the people to know about synthetic biology. Appropriate knowledge frees people from bias and equips them with discretion. In general, correct knowledge make people to be free from irrational fear toward genetic engineering. As high school students who have just entered this field, we felt that is would be important to publicize synthetic biology. Of course synthetic biology is a relatively new field in biology and is rarely introduced in high school biology classes.
- We believe that as much as synthetic biology appeals to people, it is more likely that biotechnology in whole will be accepted by the public. People avoids buying GMO and prefers organic foods. Some feel that that biotechnology in general is about 'manipulating' DNA. We thought if we tell very young students that such technologies have more potential than danger, it might change people's stereotypes.
- We created a brochure for middle school students and had synthetic biology class in NGO.
Creating a introducing brochure about synthetic biology
- Although synthetic biology is a fascinating field, it is not well known to the public. Especially in Korea, there aren’t many ways in which young students can get to know synthetic biology. There aren’t many references nor materials that can be easily read by students. So most of the students lack knowledge about synthetic biology – some may even not know what that really is. Since we hoped to share our understanding of the synthetic biology work, we made a small pamphlet that explains DNA, synthetic biology, and iGEM. We wish it would be a helpful material to all those who wish to introduce synthetic biology to everyone!
- We expect these simple, easily explained pamphlets to let students understand basic of the synthetic biology, and let people learn more about it. They might be interested in it, or feel easier when they come across the field of synthetic biology later on.
- We also introduced iGEM at the end of the brochure, too.
- The files we made is now distributed to several institutions, not only including schools but also institutions and teacher's associations.
- Feedbacks are always welcome!
- UPDATE : Here is the English Version, Too!
Synthetic Biology class in Suri Nature School
GMO quiz class
- Suri Nature School located in Gunpo, Korea, is a NGO that runs educational programs for teens. CSIA_SouthKorea participated as mentoring students who discussed students with subjects related to biology and genetic technology.
- A student in Suri nature school wanted to learn about GMO, so this is the worksheet designed to solve any basic questions regarding GMO.
- We made this quiz to grasp some basic notions about GMO & to let people know that there are some questions in genetic engineering that cannot be clearly answered "Yes" or "No"
- We made several students in Suri Nature school to solve the quizzes, and some of them were surprised by the fact that DNA is so long and it is digested in the intestines after taken by the body.
Extracting DNA from Broccoli
Brainstorming
Quorum sensing using V.harveyi
- During the project, we found that luxR part was inconsistent so that we had to have a backup plan. Groups of V. harveyi bacteria is known to communicate via quorum sensing, so we studied about its system. However, V. harveyi lacks a LuxI/R quorum sensing system, and instead employs a hybrid quorum-sensing circuit using AI-2.
- However, since AI-2 is both made by gram-negative and gram-positive bacteria, it was impossible for us to adopt part that is similar to luxR in V.harveyi. In our model of luxI/R quorum sensing system, luxI from V.fischeri (a gram-negative bacteria) only produces AHL, while aiia from B.subtilis (a gram-positive bacteria) only decomposes AHL.
- We concluded that the difference in composition and degradation rate of AHL drives synchronized oscillation. Since the system using AI-2 will not be able to provide accurate information about composition and degradation rate of AI-2, we decided not to use V.harveyi system.
Ideas from imagination
To Kill a Mosquito
- From science news articles linked below, we have found some chemicals that either attracts mosquito or kills it.
- http://www.ars.usda.gov/is/pr/2010/100309.htm
- http://www.scidev.net/en/news/mosquitoes-kill-their-own-in-new-dengue-control-me.html
- We found that ammonia, carbon dioxide, lactic acid, octenol are the substances that attract a mosquito. (Pyriproxyfen is an insecticide which is neither lethal nor repellent to human), while geranyl acetone, citronellol, geraniol, lavonax and pyriproxifen are lethal substances to dengue mosquitoes(http://www.ncbi.nlm.nih.gov/pubmed/20127888)
- Pyriproxifen works as a juvenile hormone analogue and prevents developmental steps of a larvae.
- Since a mosquito needs both its breeding area and resting place which are centered around stagnant water(toilet tanks, sinks....), it would be preferable for the lethal substance to be soluble in water.
- Professor and advisors' comments:
-Although there is a way to synthesize pyriproxyfen in a chemical way, there is no known biosynthesis pathway of the substance. -Many of the mosquito-attracting chemicals also appeal to different types of insects -Some of the mosquito-attracting chemicals are already in sales
Bacterial Sunscreen
- Bacterial production of sunscreening materials has been a recent issue in microbiology.
- We found that most sunscreens could shield UV B (280-300nm) but not UV A. In addition, most sunscreens are found to have metal oxides in their components, and the effects those could lead to melanomas. We also searched about scytonemin, a pigment found in cyanobacteria which protects them from UV radiation. Such preventative mechanisms by cyanobacterias include systems that detoxify radical oxygens produced during UV stress by using enzymatic antioxidants.
- The professor commented:
- “Sunscreen that uses bacteria is one of the interesting topics. However, for iGEM project, this idea would be difficult to realize. My lab had once also considered to synthesize the bacteria, and if the process succeeds, it would be really beneficial also in industrial terms.”
- And he sent us the paper (also marked in reference) that made us feel amazed.
- http://www.nature.com/nrmicro/journal/v9/n11/full/nrmicro2649.html
- (which is also on online)
- The advisor commented:
- The gene used in synthesizing bacteria used in sunscreen have not been completely discovered yet. Also, the pathways of the known genes are not wholly studied to use. Furthermore, about 10 to 20 operons have to be applied to synthesize bacterial sunscreen. So, it would be very difficult and complicated to make this bacteria.
Lactose intolerance curing E. coli
We thought about the mechanism of making E. Coli to do apoptosis when there is excessive nutrient in human colon (e.g. when a lactose intolerant individual intakes dairy product) so that it prevents excessive growth of E. Coli culture and reduces lactose-intolerant symptoms, such as diarrhea.
- Mechanism step (hypothetical) 1. Sensing lactose
- We could not find an adaptable lactose-sensing gene. Therefore, it would be better to adopt the glucose sensing mechanism of Baker's yeast.
- Put in lactase making gene inside the E. Coli's DNA. Put in the spliced mRNA version (of course it would be transferred into DNA by using reverse transcriptase before putting the mRNA strand into the E. Coli's gene) of Baker's yeast's glucose-sensing gene.
- When there is lactase inside human colon, the lactase-producing gene will express and E.Coli will produce lactase.
- Lactase will decompose lactose into galactose and glucose.
- The glucose sensing gene will detect glucose.
- Mechanism step 2. Apoptosis signal
- In ‘Baker's yeast’, the gene HXT is expressed when glucose is detected. Instead of HXT, if we put the gene that induces cell death in parts registry, we predicted that the cell will apoptosis when the glucose is detected.
<ref>The EMBO Journal vol. 17 no. 9 pp.2566-2573, 1998 Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces Cerevisiae</ref>
- Things to consider :
- Since the cell should not die from the signal it sent to itself, the genetically modified E.Coli should be resistant to the apoptosis signal sent from itself. Further, the power of the signal should be controlled in its time and magnitude.
- Comments from the professor and advisors
- - I would like to know whether the fact ‘lactose-intolerant symptom is created by excessive growth of e.coli inside human intestine’ is true. It would be better if you include the related papers or references that you found during the research.
- -Because lactose is comparatively rich carbon source, E.coli will have various lactose-sensing system.
- -When the gene that leads the death of cell is inserted into E.coli, it is possible to recognize lactose as gene expression signal and kill the E.coli that is not manipulated. But it seems hard to kill the natural E.coli that is in the human body. Even if it is possible, the corresponding host will also be recognized as a target. In that case, before sufficient amount of substance is made, host will be killed by the initially-generated ones. We need to think of another strategy.
Cholesterol Degradation
- This idea derived from an imagination to divide the macromolecular oil into several parts, as illustrated above. We primarily focused on our research of steroid compound degradation and enzyme that activates the process.
- The literature search was done on a very basic level. Pseudomonas sp. NCIB 10590 & Bacillus subtilis are able to degrade cholesterol to lower level. In human body, DHCR7 gene codes enzyme 7-dehydrocholesterol reductase. Gordonia cholesterolivorans (e.g., G. sihwensis, G. hydrophobica, G. australis, and G. neofelifaecis) has genes that codes cholesterol oxidase.
- Comments from the professor and advisors
- -Excess cholesterol may create various diseases, it is still true that cholesterol is essential in many biological processes. Breaking down cholesterol when its concentration is high would be desirable, but it is not that unique.
Ideas based on previous iGEM teams
Glowing Bacteria
We came up with the idea of glowing bacteria to use it as a reading lamp. Our school always shut all the electricity on 1:00 a.m. so we have to go to sleep. So, even though when we have important homework or tests coming up, we cannot study longer than 1:00 a.m. Because of this uncomfortable system, we thought of glowing bacteria that can glow without electricity so that we can stay up late and study more(really?).
* bioluminescence
- While searching for glowing bacteria, we found out that a Netherland company, Philips electronics, developed glowing bacteria that is fed with methane and produces luciferase to glow. However, they had a limitation: they said that their bacteria produces low-intensity light that it cannot be used as reading lamp. ☹
- We found another project of making glowing bacteria. This was done by Cambridge in 2010 for iGEM. They didn’t use GFP as a glowing material. Instead, they used v. fischeri lux operon. What they did was by using long-range PCR, they extracted luxCD, luxAB, and luxE individually and assembled them into new operon. And they used Gibson Assembly to make operon consisting Lux C, D, A, B, E under the arabinose-induced promoter pBAD which can activate without the gene regulator, LuxR and AHL. Also, they had made h-ns mutants that produce much brighter light than wild type strain. This gave us a hope to make a reading lamp using glowing bacteria!
- About the first ideas of making a bioluminescence lamp, the professor and advisors said that we might need a mechanical sensor.
Cobalt Buster
Team Lyon-INSA-ENS proceeded “Cobalt Buster” project about creating a bioremediation system of using Escherichia Coli biofilm to filter radioactive cobalt from contaminated water from nuclear reactors. Reflecting the fact that the formation of E. Coli biofilm is mainly caused by the production of curli, they worked on overproduction of curli, which is a highly adhesive amyloid protein.
- Understanding of the project
- They used two different approaches on doing this: one is a completely synthetic approach of creating an independent curli synthesizing pathway, and the other is activating the existing curli synthesis pathway by cloning the superactivator ompR234 gene.
- In addition, they tried to improve their strain by making their strain auxotrophic, which therefore prevent dispersion of the strain, and by inserting the transporter gene directly in the efflux pump gene. They used “Quick & Easy E. Coli Gene Deletion Kit” to delete a gene of amino acid biosynthesis and made their strain unable to survive without a medium that contains amino acid. Also, accounting that the transporter features are located on a plasmid that may not be stable and a Kanamycine resistant gene in rcnA gene knocks out the rcnA gene, they inserted the transporter feature in the rcnA gene.
- They insist that they succeeded in capturing up to 85% of the radioactive cobalt, which seems to be a successful result. Nevertheless, there are also some parts that must be improved in order to be able to be used in the real-life nuclear reactors. The temperature of the primary circuit can rise up to 327 degree Celsius; however, the adequate temperature of their biofilm is between 20 degree Celsius and 45 degree Celsius. This means that there must be four to five hours of interval before opening the reactor and passing the contaminated water through the filter, which inevitably causes economic inefficiency. In addition, in primary circuits, cobalt may exist both as ions and as particle; however, the scope of this project is limited to capturing cobalt ions and could not yet reach the ability to capture cobalt particles.
- Improvements
- Being highly interested in this project, our team thought of some ways to make significant improvements on the “Cobalt Buster” project. First, in order to improve the heat resistance of the E. Coli biofilm, we thought of adding clpK gene, which is known to “render an otherwise sensitive E. Coli strain resistant to lethal heat shock”.
- More specific plans and idea regarding the capture of cobalt particles are soon to be considered and updated.
- Comments from professor and advisors
- In high temperature, the protein is denatured because its three dimensional orientation is malformed. Therefore, it is not feasible to prevent denaturation by just inserting heat resisting gene in the bacteria.
- The characteristics of the filter were already sufficiently discovered by the previous iGEM team. So there was not much room for improvement.
- Rather, it would be interesting to study about the E.Coli's inducing mechanism against influx of Cobalt. Since bacterias are sensitive to the small intake of heavy metal compared to the influx of glucose or other essential materials, it would readily expel the ion from its internal environment. This means that present of Cobalt will greatly influence E.Coli because it is fatal. Further, it is likely that such pathways have been already discovered.
Plastic Degradation
Based on the openwetware page and Team Stanford’s brainstorming ideas(http://openwetware.org/wiki/IGEM:Stanford/2009/Plastic_Degradation#Project_Summary), we researched on phenol degradation.
- The idea of environment-friendly approach fascinated us despite difficulty of realizing the goals. We followed the ‘subprojects’ ideas on the page and tried to understand contents. If the goal of the project is to Engineer E. coli to metabolize phenol as a carbon source, linking it to cellular respiration,
- Pseudomonas sp
- http://www.ncbi.nlm.nih.gov/pmc/articles/PMC383114/
- or
- Cryptanaerobacter phenolicus
- http://ijs.sgmjournals.org/content/55/1/245
- or
- Rhodococcus phenolicus
- http://www.sciencedirect.com/science/article/pii/S0723202005000986
- should be available. However, methods of transducing bacterial plasmids into vectors that will be put in E.Coli or yeast should be investigated for further research.
However, if we use Pseudomonas sp. as the plasmid, we would have to add Na2-succinate, which is the typical source for the strain. (The related thesis was actually about the ‘simultaneous’ Degradation of Atrazine and Phenol). Cryptanaerobacter phenolicus is known as a bacterium species that produces benzoate from phenol via 4-hydroxybenzoateRhodococcus.
Synthesis of Flavor using Candida Rugosa
- Our extensive research about lipase led to investigation of some specific function of the lipase, including flavor synthesis. The thesis linked below revealed us that C.rugosa could synthesize pentyl propanoate, isopentyl butanoate, and butyl butanoate, which are all components of apple flavor, by using lipase.
- Based on the source from openwetware(which is also a project for MIT in 2006),
- we thought of a device generating apple flavor. In the project in openwetware, the promotor codes ATF1 enzyme, which converts isoamyl alcohol to isoamyl acetate. If we find specific lipase that makes pentyl propanoate or isopentyl butanoate , and leave it in adequate substrate(we should research more!!) we thought that the synthesis of flavor would be feasible.
- The professor first commented that it would be good to quantitatively measure the amount of fragrance(gas), but he said that gas chromatography would be unavailable and complicated. We later agreed to reserve the idea of quantitative measurement of gas.
- We found out that 2006 MIT iGEM team did not either quantitatively measured the strength of fragrance. They applied relative arbitrary scale in their measurements.
- Comments from professor and advisors
- This type of project was widely studied by numerous previous iGEM teams. So, it would be repetitive to address the same task.
Methane Sensing
The team thought of developing methane sensing devices based on project of METU, Turkey.
- Understanding of the project
- They had four steps in implementing their project :
- Methane sensing(in fact, MMO sensing), conversion of MMO into methanol, entrapment of methanol using enzymes, and a killswitch(cessation of replicating the sensor). The team found that the bacteria ‘Pseudomonas oleovorans’ find alkane and degrade it for carbon source.
- However, according to the team,
- -the synthesized methane monoxygenase(MMO) construct was such a long part. The main methane interacting region of monooxygenase could not be expressed functionally.
- Improvements
- We thought of improving the first step of METU’s project, by more thoroughly investigating about the activator protein AlkS. AlkS induces the transcription from PalkB promoter which initiates the expression of genes code for assimilation of alkanes.
- Still, we were assured by the following informations:
- -sensitive to methane presence and have mechanisms to activate transcription of related gene clusters.
- -the transcription is expressed in E.coli correctly.
- Comments from the professor and advisors
- Since MMO is a multimeric enzyme, it would be difficult to be fully expressed. (This means that the problem that was in the original project could not be easily solved)
Diary
- Please see our Diary.
Sep~Dec 2011
- 09.20.2011
- -We didn’t know any professional knowledge about synthetic biology so we decided to ask for help of experts. While searching for the professors who participated in iGEM before, we found out that a professor from Korea University has participated several times before. So, we first sent an e-mail to our instructor In-Geol Choi to ask for his help.
- 09.21.2011
- -The professor responded very quickly. And he was really positive toward us having interest in synthetic biology and willing to participate in iGEM.
- 12.18.2011
- -While searching through the iGEM wikis, we got interested in making bacterial sunscreen, the idea from the imperial college 2011 wiki brainstorming section. Sunscreens that we normally use can only protect UVB, which is only one part of the broad range of UV. So, we were hoping to make bacteria that can protect UVB and UVA, another kind of UV light. And according to our further research about bacterial sunscreens, scytonemin, a pigment synthesized in cyanobacteria, absorbs UVA and pityriacitrin, a pigment synthesized in malassezia yeast, absorbs a wide range of UV. Thus, we thought we could use the genes that codes for this pigments and make a bacteria that carries the genes. However, we had so much difficulty finding it that we sent the second e-mail today for help from the professor.
- 12.19.2011
- -We received an answer from our instructor. He said our idea was interesting and worth making it, but it would be too hard for us to make it as high school students. ☹ He also sent us a review related to bacterial sunscreens from the Nature: Microbial ultraviolet sunscreens by Qunjie Gao and Ferran Garcia-Pichel.
- 12.25.2011
- -We sent our first e-mail to iGEM asking several questions about the competition.
- 12.28.2011
- -We received a reply from the iGEM and she was very nice! ☺
Jan~Feb 2012
- 01.13.2012
- -Today, we sent another e-mail to our instructor with several ideas. All of these ideas were from previous team wikis. From open wet ware, we found the rough outline of Stanford 2009 iGEM project. This team tried to degrade plastic, specifically phenol and formaldehyde, into carbon source of e.coli. However, we couldn’t find any sign that shows the team actually conducted the experimentation. Other one was making an e.coli that can sense methane. This idea was based on METU 2011 project. This team, had several projects-sensing methane, conversion, entrapment, killswitch- but they didn’t accomplish their goal. So we thought we could try part of their project, methane sensing. The last idea we sent was making e.coli that produces an apple smell. We found a similar project, MIT 2006 project, in biobuilder homepage that was to make a banana odor generator with e.coli.
- -More information about our ideas can be found in brainstorming section of our wiki!
- 01.13.2012
- -We received some negative comments on our ideas. He told us that since methane oxygenase is multimeric enzyme, we would have hard time sensing methane and also plastic degrading would be a tough task for us. But he said maybe using other kind of ester to produce odor could be possible. And he also suggested us to improve Korea University 2010 project.
- 02.16.2012: meeting with professor
- -Our team had a meeting with professor In-Geol Choi in order to discuss our topic for the igem project. We had some creative ideas of our own, but we weren’t sure if they were appropriate topics for this project. As our discussion went on, professor Choi gave us some advices in selecting the topic:
- -First, the project should have some kind of purpose – something like ‘saving environment’. We need to synthesize and create biological machine of certain function that can fulfill the fundamental purpose of our own.
- - Second, considering that we don’t have much time nor the professional knowledge for the research and experiments, as high school students, it might be more appropriate to work with relatively easy project. He recommended us to do some research on websites such as biobuilders, and read about some labs that students can conduct.
- -Third, note the parts registry that we can use. In parts registry site ( http://partsregistry.org/Catalog) we can find out which devices and functions we are able to use, and that will help us with selecting our topic. We decided to consider these points and brainstorm more about the topics. We are to determine our topic by the end of this February.
- 02.20.2012: ideas, ideas, ideas
- -We first start to have an idea of making glowing bacteria to use it as a reading lamp. And having done some research, we found out that most of the glowing products were containing a protein called GFP (green fluorescent protein). To make a new challenge, we thought of making a new color of light other than green light. However, we soon found out that other great scientist have already made numerous mutation of GFP and produced many other colors. So, now… we are stuck!!!
- 02.17.2012
- -After some discussions and research, we came up with some ideas: Lacotse intolerance curing ecoli, cholesterol degradation, and synthesis of scent. We referred to many of the previous igem projects and the research materials to determine necessary steps for each of the project.
- -Lactose intolerance curing e.coli : mechanism of making E.Coli to do apoptosis when ther is excessive nutrient in human colon
- It prevents excessive growth of E.Coli culture and reduces lactose-intolerant sypmtoms
- 1) Sensing lactose
- 2) Apoptosis signal
- -because cell can’t die due to its own signal, genetically engineered E.Coli should be made resistant to apoptosis signal
- -To prevent complete eradication of E.coli or too minimal deaths of E.Coli, the strength or period of signal should be controlled
- -Cholesterol degradation
- Bacteria called Pseudomonas sp. NCIB 10590 and Bcillus subtillis have the gene to degrade cholesterol. In human bodies, DHCR7 gene codes for enzyme 7-dehydrocholesterol reductase.
- -Synthesis of scent using Candida rugose
- (*openwetware)
Mar 2012
- 03.05.2012
- -The idea of synthesis of scent
- When we looked over the banana scent-synthesing project that MIT had conducted before, we found out that they applied arbitrary standard for the quantity of scent. But other than that, we couldn’t find a method of quantitatively measuring specific gas. We are still looking for mechanical sensors..
- 03.07.2012
- We thought of additional ideas: killing mosquitoes and toggle switch.
- -Killing mosquitoes
- Mosquitoes have a preference for dextorotatory ocetenol molecules that are emitted y mammals.
During the research, we found out how to lure the mosquitoes and chemicals that can kill them. Engineering E.Coli that can synthesize both chemicals would be the goal.
- -Toggle switch
- Two positive feeback loop that represses each other
- Idea 1) make an input that can sensor pH, and distribute in in the soil so that weak base can be released when needed
- Idea 2) because toggle switch has similar mechanism with neuron, it might be able to model a simple neuron circuit.
- 03.12.2012
- We received some negative feedbacks for our “cobalt buster” proposal. We made this proposal based on previous igem projects. However, he told us that it doesn’t seem to be able to be significantly improved, nor be more developed, since previous igem team had already completed much of the work. Instead, what we should focus on is developing the projects that prvious igem team had failed. What he advised was that because there is not enough time for highschool-division to go over all the errors and fix them, it would be better to plan a project with interesting ideas. He also suggested that in Cobalt Buster project, we can modify the operon so that we can create a circuit that has cobalt as the inducer.
- 03.13.2012
- Professor Choi also sent us some feedbacks on our ideas. He first commented on our “mosquito-kiling” that it is a great idea, but we should consider more about the feasibility, and how long it would take to complete the project.
- Also, he told us to do some more research, comparing toggle switch and repressilator. (http://en.wikipedia.org/wiki/Repressilator)
- Also, we decided to make lists of our ideas and decide one among those by the next week.
- 03.14.2012
- Another feedback arrived:
- 1. Pyriproxyfen, the substance that kills mosquito work as juvenile hormone analogue to prevent larvae’s development. However, we need a way to synthesize this substance other than in chemical way.
- 2. Toggle switch seems to be a similar system with the repressilator. It would be interesting to create microorganism with circadian rhythm or biological clock. We will discuss more about how to use it.
- 3. Cobalt buster seems to be unfeasible only with a few heat-resistant genes.
- 03.16.2012
- We did some research about what our professor wanted us to do about toggle switch. Toggle switch is a similar system with repressilator, and the only difference is that toggle switch has two loops repressing each other and repressilator has three. And we thought circadian rhythm idea was really great.
- Also, we developed our previous idea of making glowing bacteria using mechanical sensors. We thought of making a grid with 10*10 or more spaces, place gfp, and let each space have signal of 0 or 1, controlling the coloring of the bacteria in each space. It would be sort of like making the bacteria-neon sign.
Then we got reply from the professor. He told us that mechanical sensor would be found if we search previous iGEMs. For the grid idea, it would be possible with e.coli with toggle switch and turn it on or off.
- 03.17.2012
- (comments from professor & advisors)
- 1. MIT team in 2010 iGEM made a system with mammalian cell that responds to the pressure. It is uncertain whether we can apply eukaryotic promoter to the prokaryote or not. Also, it wouldn’t be so easy to find a system in which microorganism responds to the physical stimulus. Instead, it might be possible to use ‘heat-sensing’ property, shaking it to raise the temperature and making it respond.
https://2010.igem.org/files/poster/MIT.pdf (MIT 2012 iGEM poster)
- 2. To develop more about the idea of coding the grid system with toggle switch, we can use the fluorescent-protein as a reporter for the switch. When switch is off, it will have value of 0 and 1 when it is on. We can use filter paper and put only on some of them the inducer that turns the switch on, and place it on e.coli. Then, it will have fluorescent-protein appearing on only some of filter papers, and when we send it, receiver can use the wave that excites the protein to get the message.
- We will be meeting with the professor next Wednesday (3/21) and make final decision on our topic for the iGEM project ☺
- 03.18. 2012
- Professor Choi commented that Electronic display using bacteria seems to be the best project for us to experiment.
- He said that “The example of the code below is the “on only” experiment which doesn’t need toggle switch to turn it on and off.
- The other possible suggestion is that first, make the circuit that works with toggle switch, and in the 96 well plate, control on-off of the each well and make the fluorescence oscillate (on and off). Then it would seem like electronic display.
- I will discuss it with assistant and tell you whether it is possible or not. You can talk with him on Wednesday and make final decision.”
- 03.19.2012
- We organized our brainstormed ideas before we have a meeting on Wednesday:
- Sunblock utilizing bacteria
- Resisting mosquitoes
- Electronic display using bacteria (and toggle switch)
- cobalt detection (modifying the operon in order to create a circuit that has cobalt as the inducer)
- glowing bacteria with heat sensor
- lactose intolerance
- cholesterol degradation
- glowing bacteria
- plastic degradation
- synthesis of flavor
- methane sensing
- We are also organizing wiki with the ideas that came up till now.
- 03.23.2012
- First, we made decision on our research project! We are planning to work on bacteria-display.
- We also uploaded wiki with what we discussed with the assistant on Wednesday.
- Professor Choi commented that-
- Substance that allures mosquitoes applies not only to mosquitoes but to all the insects, so it won’t work specifically to the mosquitoes.
- Biosynthetic pathway isn’t well known, and the substance itself is already commercialized in the market, so it doesn’t seem to have much meaning.
- Also, 1. Try making team logo 2. Upload team introduction to wiki.
- 03.25.2012
- We asked several questions.
- 1. 96 well plate has a lot of wells each, and we are curious how gfp in separate wells uses quorum sensing to synchronize their oscillation periods.
- 2. You told us that the period that fluorescence appears and disappears is the same, so that we should use a video; we are curious how long it goes!
- 03.27.2012
- Instructor told us that our logo seems to be simple and nice^^ and added that it would also be nice to make a logo that represents the project. He also told us that for iGEMHS, the part registration is not required; igem homepage says that special awards are given to teams who register parts with “new, cool and important function”
- And here are the answers:
- 1. Synchronization of gfp expression in one well is explained in the paper linked below.
- In Fig1(a) diagram, luxI promoter that is originally used in quorum sensing can be modified to function as repressilator.
- http://www.nature.com/nature/journal/v463/n7279/full/nature08753.html
- 2. According to the paper, repressilator has about 150 minutes long period, and quorum sensing about 70 minutes. But it can differ according to the laboratory environment, so we will have to measure it ourselves during the experiment.
- We should first search the parts related to repressilator or toggle switch, and see if we can utilize it.
- 03.28.2012
- We didn’t really understand why quorum sensing has 70 minutes of period, so we asked the instructor.
- In fact, reading the paper was an excruciating process but the result was worth it.
- 1. If we use E.coli with shorter period then we can reduce the time; does it mean that the period can be shorter than 70 minutes?
- 2. We found a picture for repressilator, but it says that c gene at the end represses gfp, so we aren’t really sure what’s right…
- 3. We are looking up parts registry site to find gene that goes into repressilator, but we are not sure what gene is needed☹ Are we using the same genes that Elowitz and Leibler used?
- 03.29.2012
- Instructor sent us two papers, the one with Elowitz and Leibler & the one that is linked above.
- He also told us that reducing the oscillation period shorter should be considered after we check that the system works properly. He said: “About gfp repression, you will know better if you read the paper. Based on the picture that you sent me, TetR gene is repressing the gfp expression, and gfp is expressed when λ cI gene is expressed. Also, I think that it would be too hard to create a new promoter and make oscillation system. It would be both easier and much certain to use the same method as Elowitz and Leibler’s. If you search ‘igem repressilator’ on google, you’ll see that there were many previous igem teams who used the same circuit. Laboratory schedule isn’t decided specifically. I’ll tell you as soon as I can after I discuss it with the professor.”
- 03.30.2012
- We asked several questions again!
- 1. Both two papers are about gfp oscillation occurring in one colony; I’m curious whether we have to connect every single cells in 96 well plate into one colony
- 2. When doing the lab, is the trapping chamber necessary for making critical cell density as shown in 2010 paper? If we do so in 96 well plate, then how can we control flow rate?
- 3. In 2000 paper is the IPTG inducer that has to be inserted from the external environment? And moving the cell from place with IPTG to without IPTG resulted in the occurrence of gfp fluorescence oscillation?
On the other hand, AHL is substance synthesized by luxI, so amount of AHL also oscillated in the 2010 paper? Then I’m curious that if we have to use autoinducer similar to AHL.
- 4. I also want to ask if substance that is used to do cell-to-cell coupling wasn’t used in 2000 paper. We haven’t been able to read closely the previous igem projects yet. Always thank you for kind reply!
Apr 2012
- 04.02.2012
- We kept reading a thesis by Elowitz & Leibler.
- We were curious about why the oscillation slows down suddenly at the post-septation phase. (Then, doesn’t the oscillation happen in the E. coli after about 600 minutes, as shown in the graph?) Further, it was hard to understand why only about 40% of the cells exhibits oscillation behavior. In the thesis, it is stated that transient pulse of IPTG might be capable of synchronizing, because IPTG hinders the repression of LacI. We do not understand why. We asked these questions to professor Choi.
- 04.03.2012
- This is the reply from professor Choi☺
- According to the thesis, when the cell (E. coli) enters the stationary phase, the oscillation stops. (p.335 first paragraph, last sentence). The reason for only 40% of the cells showing the oscillatory characteristic may vary. Fundamentally, the oscillation is the result of complex cellular metabolism, not simple chemical reactions, so the oscillation may be highly affected by various external circumstances, such as the size of the cell, metabolism rate, diffusion rate, etc. So, as the time that the cell is exposed to IPTG becomes longer, synchronization will be hindered more, due to the same reason. I think making the circuit is not very difficult, but obtaining preferred results will not be a easy thing. We’ll make up the schedule to proceed experiments for building the circuits with your instructor.
- 04.05.2012
- We thought about make oscillation of GFP expression in one colony in a 96-well-plate and size of the colony. Instructor told us that the thesis about the repressilator is about oscillation in a single E. coli cell, and the other thesis is about synchronization of oscillations in a cell colony, not a single cell. In our experiment, we will use the 96-well-plate to raise the cells in different wells as independent colonies. We will not make the whole plate as a single colony.
- We were also curious about use of ‘trapping chamber’ in order to create the critical cell density. Our experiment will be conducted inside a 96-well-plate; we wonder how we could control the flow rate inside the plate, if we use the same mechanism as the ‘trapping chamber’ in order to obtain the critical cell density. We concluded that it will be quite difficult to use a trapping chamber to make a continuous culture. Instead, we will use batch culture system to raise the cells.
- We asked our instructor whether we understood the theses correctly: is IPTG an inducer that must be injected from external source? And because AHL is a substance that luxI synthesizes, the amount of AHL oscillates together, right? Then, if we use quorum sensing in the experiment, do we need to use autoinducer, such as AHL?
He replied that IPTG is a chemically synthesized substance, and it is to be injected from external source. AHL is auto-synthesized. Therefore, the concentration of AHL also shows oscillation, and we do not need to put in AHL.
- We also discussed that due to the difference in the period of oscillations among the cells, preferred results may not come out in our experiment. But, because repressilator parts are already registered in the parts registry by previous iGEM teams, the experiment is relatively easy to conduct right now. Instructor said that we will start preparing for the experiment and order the parts from the part registry, so we should design a theoretical model of our experiment to see if our experiment is actually plausible.
- 04.06.2012
Designing the experiment was quite a difficult process than we thought. So we asked professor for some help: 1) Is there any method to regulate the difference of oscillation period among individual cells? (Expect for putting in IPTG at the same moment)
2) If we conduct the experiment with a single cell, will the GFP expression be bright enough to be seen with naked eye?
3) Is doing the quorum sensing more difficult than building the repressilator circuit?
- 04.11.2012
- We got answers to several questions that we asked the professor.
- 1) We can start the laboratory from the third week of April. First, we’ll make oscillation using repressilator or quorum sensing mechanism.
- 2) The instructor will be preparing the experiment plan, and doing the experiment approximately once a week will be appropriate.
- 3) If we conduct the experiment with a single cell, a fluorescence microscope is needed to observe the expression of GFP.
- 4) The circuits are not difficult to build, if all the parts are available. (but the circuit’s viability is a different issue)
- 5) The synchronization of oscillation seems to be difficult, but we will try.
- 6) If possible, the team members should do an in silico model experiment, using the differential equations stated in the thesis. Through the modeling, determine the parameters that the amplitude and frequency of the oscillation can be differed.
- 04.13.2012
- However, due to pending AP exams that we have to take, we won’t be able to attend the lab from last week of April to the third week of May.
- Also, we are going to try studying the differential equations.
- 04.25.2012
- We again asked some questions on the model that we are going to use.
- 1. What is the reason for a repeated use of luxPL and luxPR promoters?
- 2. What is hybrid promoter? (It says that the promoter I751502 in front of luxI is a hybrid promoter.)
- 3. I wonder if the luxR-AHL complex is readily degraded. Is the degradation rate similar to that of AHL?
- 4. Can you explain more specifically about the protocol?
- 04.26.2012
- E-mail from the instructor:
- Instructor sent us e-mail with the answers to our questions:
- 1) luxPL and luxPR are different promoters with different characteristics. (luxPL: a constitutive promoter, lux PR: a promoter inducible by AHL-luxR complex) For luxR protein, because the protein needs to be continuously expressed, luxPL promoter is used, and for GRP, luxI, and Aiia proteins, oscillations in the protein concentration is required, so luxPR is used.
- 2) Hybrid promoter is not a component that we will use in our experiment. I1751502 is a combination of luxPR promoter and lac operator site; it maintains the original characteristic of luxPR (inducible by AHL-luxR complex) and has lactose-inducible characteristic at the same time.
- 3) Both luxR-AHL complex and free AHL degrade at a similar rate with that of Aiia. When both proteins are degraded, GFP expression turns off.
- 4) The standard protocol of cloning will be easier for you to understand if you read the explanation from the iGEM homepage.
- 04.29.2012
- Oscillation model wasn’t an easy thing to understand completely; we had endless questions! We explained some parts that we understood, and then asked the instructor and professor what we didn’t understand yet.
- As the luxI promoter becomes stronger and the amount of AHL becomes greater, the reaction rate increases (because of increased amount of luxR-AHL complex), and therefore, it is expected that the oscillation period will be shorter.
- Then, 1) Is it also predictable that more GFP will be expressed as well?
- 2) Is it possible to predict its amount quantitatively?
- In the wiki of Team Wageningen, it is stated that due to the tendency of interconnected positive and negative feedback loops to reach a steady state rather than sustaining oscillations, the external conditions need to be precisely controlled in order for the system to produce synchronized oscillations. Then, 1) How can we maintain the cell density for a long period of time?
- 2) Or, is it unnecessary to artificially maintain the cell density, because quorum sensing continuously occurs after certain threshold density?
- 04.30.2012
- Professor e-mailed us again and told us that 3A assembly kit will be arriving this week, so that we can use it in the experiment (http://partsregistry.org/Help/3A_Assembly_Kit) : Also, the calendar of events can be looked up in the link shown, and we had to keep track of schedule that we have to go through. ( https://2012hs.igem.org/Calendar_of_Events) And the judging criteria can be found here ( https://2012hs.igem.org/Judging).
- We now need to decide the time for our experiment and start working on it. Instructor will help us throughout the experiment, and he will be including the experiment for observing the growth curve of E Coli and measuring/observing fluorescent proteins.
May 2012
- 05.05.2012
- We learned about biobrick standard assembly and 3 antibiotic (3A) assembly. Standard assembly is a two way ligation with two biobricks and a vector. 3A ligation is a three way ligation using three biobricks, vector, and three antibiotics to get only the successfully transformed cell. When each biobrick part is cut with different enzymes, they possess sticky ends that enable them to combine. However, once they are combined, they make mix-ends which would not be recognized by any of the restriction enzymes.
http://partsregistry.org/Assembly:Standard_assembly
- We went over biobrick standard assembly / 3A assembly
3A assembly relies on three way ligation for assembly rather than two way ligation between a part and a part + vector molecule(standard assembly) different sticky ends are able together even if they are cut with different enzymes but such part become mixed-ends, which means they would not be recognized by any of the restriction enzymes.
- 05.10.2012
- We further studied assembly methods in the website below:
- http://www.bio.davidson.edu/courses/molbio/BioBricks/BioBricks.html
- According to this website, all restriction sites are palindromes, top strand 5’ to 3’ being identical to the bottom strand 5’ to 3’, which is called sticky ends. Mixed sites are not palindrome, so they cannot be cut by any restriction enzyme.
- 05.12.2012
- We learned about what is a linearized plasmid backbone. By using linearized plasmid backbone, the remnant cut with EcoRI and PstI is short that it will not ligate. And this allows to build high quality construction plasmid backbone without purifying the cut fragments remaining after PCR.
2012 DNA Distribution came with a set of linearized plasmid backbones: pSB1A3, pSB1C3, pSB1K3.m1, and pSB1T3.
- 05.21.2012
- We further studied about protein purification using gel electrophoresis in standard assembly. Protein purification is series of processes intended to isolate a single type of protein from a complex mixture. pH graded gel / ion exchange column / size exclusion chromatography / SDS-PAGE are examples.
- 05.22.2012
- We read an article about the protocol from iGEM 2011 Team Wageningen wiki, and wondered if there is any process from the protocol of Team Wageningen that we are not planning to use.
- Also, we could find many kinds of protocols, other than the one that Team Wageningen has used, such as:
· Preparing chemically competent cells · Preparing TSS buffer · Transforming chemically competent cells · Preparing chemically competent cells (Inoue) · Transforming chemically competent cells (Inoue) · TOP10 chemically competent cells
- So we asked the professor what we are planning to use among them.
- Also, there are some questions about the article written by Team Wageningen:
- 1) What does OD stands for?
- In Wikipedia, it is indicated that OD stands for either ‘optical depth’ or ‘optical density’, but we cannot figure out in what meaning was ‘OD’ used in the protocol.
- 2) What does ‘OD 600nm of 0.3’ mean?
- 3) And, why should the OD value changed into 1.0~1.5 later on. What influence does OD have in the experiment?
- 05.23.2012
- And our instructor gave us the answers for our questions☺
- He told us that OD stands for ‘optical density’. The OD value is proportional to bacterial cell number. The cell number is larger if the OD value is larger.
- From the OD value, we can predict not only the number but also cell condition. During the phase in which cells grow actively, the OD value increases continuously, but after certain period of time, the OD value does not increase any more.
- In this phase, the cells stop cell division and get themselves prepared for death.
- When we conduct the experiment, it is better for us to do it with healthy, young cells, rather than old, weak cells.
- Therefore, in the protocol, the OD number is controlled to regulate the cell number or to use cells in good conditions.
- Also, he asked us when we could visit the lab. And if we have time, he is going to tell us the experiment protocol starting from making chemically competent cells. He told us that he is currently working on the cloning of the cells.
- 05.30.2012
- Our team member Ju Yeon Han and our instructor Hong Jae Park ( participated in the iGEM online meeting at Second Life. During the meeting, we could talk to the iGEM headquarter and another iGEM high school team from Mexico. At first, we talked about what we experienced through iGEM; our team member said that she learned a lot from reading scientific papers and learning more about synthetic biology that we can barely learn from school. The participants of the online meeting also exchanged ideas about socializing at the jamboree; some ideas were suggested, such as running a café, Xbox game room or a basketball court.
- Then, Ju Yeon asked a question about organizing the wiki; igemHQ told her a way to create new pages in the wiki. Our instructor also asked a question; it was about a problem in the assembly process. Our instructor tried to assemble the parts using the 3A assembly method, but unexpected results kept coming out; for example, red colonies were found, although white colonies were expected to be made. Jimmybly (the Mexican team) was also having some problem regarding the linearization of plasmid and was having some unexpected results, such as finding white colonies when red colonies are expected to be present. Also, our instructor said that when he cut luxR, luxI, aiia and GFP from psb1A2, the vector size of luxR turns out to be unexpectedly large; he later on found out that the part that we were using in our experiment was actually inconsistent.
- In the later part of the meeting, the participants of the meeting exchanged ideas on the issue about the ‘motivation’ of participating in iGEM and some advices to improve iGEM administration.
Safety
References
- ere
<forum_subtle />
 "
"